Staphylococcus pseudintermedius: an undocumented, emerging pathogen in humans
Suneel Bhooshan 1Vikrant Negi 2
Prabhat K. Khatri 1
1 Department of Microbiology Dr. S. N. Medical College, Jodhpur, Rajasthan, India
2 Department of Microbiology, Government Medical College, Haldwani Nainital, Uttarakhand, India
Abstract
The first infections of methicillin-resistant Staphylococcus pseudintermedius in humans were recorded in 2006, and is now becoming a concern because of its close similarities to human pathogens in the Staphylococcus intermedius group (SIG). These bacteria have all the properties which a multidrug-resistant Staphylococcus aureus possesses.
The literature was searched using the term “Staphylococcus pseudintermedius” in PubMed and other reference databases. The virulence factor and the pathogenicity are under investigation, but reports have suggested that this commensal of animals is transmitted easily via close contact to animals by owners, veterinarians and staff.
Resistance to beta-lactams (including methicillin) is a primary concern. Drug resistance to methicillin is a considerable problem in developing countries, as antibiotic use is not regulated. Studies from Europe have reported multidrug resistant isolates from clinical specimens. Although data on drug resistance and pathogenesis of S. pseudintermedius are not sufficient, it is extremely important to identify the pathogen correctly. Only then can its pathogenesis be studied during the course of disease and appropriate measures developed to prevent it becoming a global problem.
Keywords
Staphylococcus aureus, Staphylococcus pseudintermedius, zoonosis
Introduction
The genus Staphylococcus is currently divided into 38 species and 17 subspecies. It is infamous for its drug resistance and multiple pathogenic factors [1]. Based on the presence of coagulase enzyme, genera were broadly divided in two categories: coagulase-positive and coagulase-negative species. Initially, only Staphylococcus aureus were thought to be a human pathogen, but in 1976. Staphylococcus intermedius, a new coagulase-positive species, was identified and reported to be associated with animal and human infections [2]. Staphylococcus intermedius was first considered to be a single species. Later, based on 16S rRNA typing, it was reclassified as Staphylococcus intermedius group (SIG), including three species: S. intermedius, Staphylococcus pseudintermedius and Staphylococcus delphini, which were closely related in terms of biochemical reactions. In this group, only S. intermedius was considered to be pathogenic in humans. S. pseudintermedius and S. delphini are canine commensals or opportunistic pathogens associated with skin and wound infections, predominately in animals. In recent veterinary literature, S. pseudintermedius is one of the important pathogens of zoonotic origin that causes wound and skin infections. According to the literature, up to 90% of healthy dogs may be colonized with S. pseudintermedius [3], [4]. S. pseudintermedius mimics S. intermedius phenotypically, which makes its identification difficult using automated identification systems. Until the last decade, it was falsely reported to be S. intermedius by phenotypic and automated systems, owing to a great paucity of data available for identification. Not all commercially available identification systems are able to correctly identify S. pseudintermedius.
The unjustified use of antimicrobials in companion animals is responsible for emerging antimicrobial resistance. S. pseudintermedius is another link in the same chain in emerging drug resistance, as it is reported to be multidrug resistant, able to transmit from animals to humans, and possesses all the virulence factors of S. aureus.
In 2006, the first cases of S. pseudintermedius infection in humans were reported by Van Hoovels [5] from 60-year-old patients with clinical presentation of ischemic cardiomyopathy and ventricle tachycardia, but it has likely been present in the community for far longer. Since then, there have been attempts to isolate and categorize this pathogen to study its virulence factors and pathogenesis in humans [5], [6]. The spectrum of infections caused by S. pseudintermedius is very close to S. aureus infections. A case series of 24 isolates by Somayaji in 2016 shows comorbidity factors, with the elderly being more prone to infection [7]. Only 2 patients (8%) out of 24 were below age 40, out of which one had a wound infection related to a dog bite.
Methods
Using the keyword “Staphylococcus pseudintermedius”, we searched PubMed, finding total of 339 publications including both veterinary and human medicine, out of which 72 were reported from humans. The search also included google and public health agency information (National institutes of health [NIH)], Centers for Disease Control and Prevention [CDC], the European Centre for Disease Prevention and Control [ECDC], the US Food and Drug Administration (FDA), Agency for Healthcare Research and Quality [AHRQ], etc). We reviewed all literature published including research articles, original articles, review articles and case reports from human and veterinary medicine through July 21, 2018. The search strategy included only English-language publications.
Results
Genetic characterization
Several molecular methods are used for differentiating S. pseudintermedius from the Staphylococcus intermedius group (SIG, S. intermedius, S. pseudintermedius, and S. delphini), but these are limited to research purposes owing to its cost and lack of clinical association with disease. Ribotyping and PFGE are some of the various DNA-based techniques are used for S. pseudintermedius typing and epidemiological surveillance [8], [9], [10], [11], [12], [13]. In recent research-based studies, PCR-RFLP, spa typing and MLST are also used for typing [14], [15], [16], [17]. MALDI-TOF MS has shown promising results in identification and differentiation of SIG, although the sensitivity and specificity are not better for S. intermedius than for S. pseudintermedius [18]. Focusing on antibiotic resistance, multiplex PCR and SCC mec gene typing have been studied for macA gene detection, which is responsible for methicillin resistance.
Biochemical identification
S. pseudintermedius must be differentiated from other coagulase-positive Staphylococcus species by using a combination of biochemical tests (Table 1 [Tab. 1]). On blood agar plate, it shows creamy white colonies with beta-hemolysis. The lack of biochemical and automation resources to differentiate between coagulase producing species of Staphylococcus group usually leads to erroneous reporting of all coagulase producing species as Staphylococcus aureus.
Table 1: Phenotypic tests for differentiation of coagulase-positive Staphylococcus species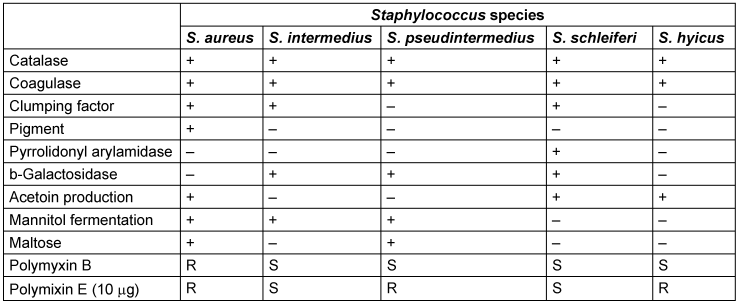
Staphylococci are grouped together as S. aureus. The arginine dihydrolase test, β-gentibiose test, D-mannitol and polymyxin B disk differentiation tests are important biochemical assays which can differentiate S. pseudintermedius from other closely related Staphylococci [5], [18], [19], [20].
Pathogenic factor and pathogenesis
Pathogenic factors are very similar to S. aureus. Knowledge about pathogenesis of S. pseudintermedius is very limited in the case of strains originating from humans. Enzymes and toxins produced by S. pseudintermedius have shown same activity in in vitro tests (Table 2 [Tab. 2]).
Table 2: Resemblance of pathogenic factors in S. aureus and S. pseudintermedius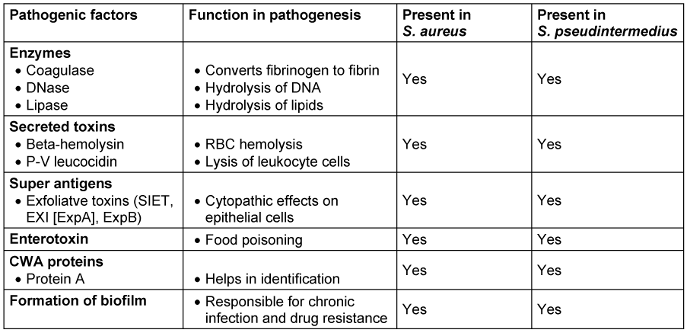
Panton-Valentine leukocidin of S. aureus is a cytotoxin that destroys leukocytes and causes tissue necrosis. A similar toxin, bio-component leukotoxin Luk-I, encoded by two genes, lukS/F, is also produced by S. pseudintermedius. Pathogenesis in humans has not been thoroughly studied and requires more detailed investigation.
S. pseudintermedius is an opportunistic pathogen. It is part of the normal flora of most dogs and does not cause any disease, unless the resistance of the host is lowered and the skin barrier altered by predisposing factors, such as atopic dermatitis, medical and surgical procedures, and or immunosuppressive disorders. Similar to S. aureus infection in humans, colonization is likely to be a risk factor for infection and, in most circumstances, dogs are likely to become infected with a strain that they carry on their body [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58]. Case reports from implant devices have yielded alarming results about its pathogenesis, which suggests biofilm formation. Staphylococcus genera are well-known for their biofilm-forming properties. Pomilio 2015 [59] conducted an interesting and novel in vitro study to demonstrate biofilm formation properties by providing a simulated environment similar to wound infection by adding serum, adjusting pH and antibiotic concentrations for 48 to 72 hours of exposure. The results of that study demonstrated the ability to form biofilm in vitro for the first time. Along with these findings other properties were also noted, such as the effect of serum and production of abundant amounts of extracellular polymeric substance (EPS) matrix, as observed by scanning electron microscope. This simulation suggested that this bacterium can produce biofilm on implant devices such as catheters, and is able to survive in wound environments by producing excessive amount of EPS, which stops antibiotic penetration in to biofilm.
Epidemiology
S. pseudintermedius was initially misdiagnosed as S. intermedius due to lack of data. Many details are still not available about epidemiology, transmission and risk factors, although on the basis of genetic linage, it has now been confirmed world-wide. It was first reported from Belgium, and later in other countries with different signatures in their genetic makeup when categorized by multilocus sequence types (MLST) and spa types (Figure 1 [Fig. 1], Table 3 [Tab. 3]). It is a part of normal flora in canines, colonizing the mouth, nose, perineum and groin. The transmission route is vertical in animals and horizontal or interspecies in the case of veterinary staff and dog owners via close contact with colonized pets. Risk factors in humans are immunosuppressed status, postsurgical infections, and old age. So far, there is no evidence of transmission of this pathogen between humans to human [20], [25], [58], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82].
Figure 1: Global demographic data (reported cases in grey)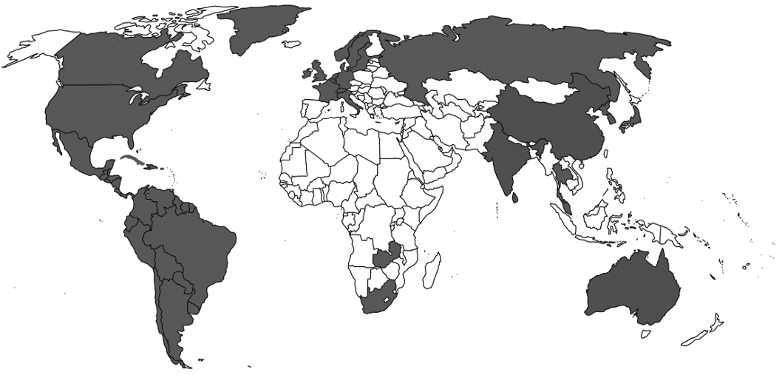
Table 3: Type of study and country reported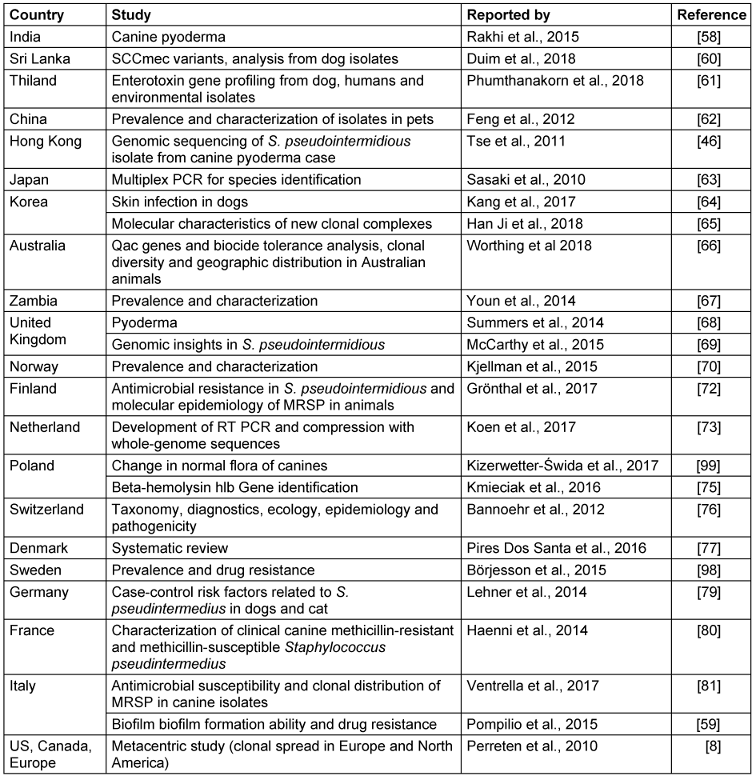
S. pseudintermedius exhibits variable clinical manifestations from superficial infection to invasive infections (Table 4 [Tab. 4]). In dogs S. pseudintermedius is mostly associated with skin and soft tissue infection, but in humans it has been reported from various sites, such as the endocardium (endocarditis), ear (otitis externa) and prosthetic joints (infections) [5], [6], [7], [19], [24], [83], [84], [85], [86].
Table 4: Clinical diseases caused in humans and dogs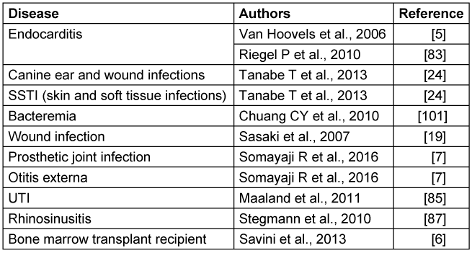
Drug resistance
In the last decade, phenotypic and automated methods have not been able to differentiate between S. intermedius and S. pseudintermedius, and the drug resistance pattern is not well studied. Thus, definitive statements cannot be made yet. However, drug resistance in S. pseudintermedius has been reported by some authors in veterinary isolates [87]. The highest resistance rates of Staphylococcus species are against beta-lactam antibiotics, with almost 95% of the clinical isolates being resistant to penicillin [88], [89], [90]. The resistance mechanism of S. pseudintermedius is the same as in S. aureus. Drug resistance to beta lactams are mediated by Staphylococcal Chromosomal Cassette (SCCmec). The mecA gene is transmitted by plasmids between different Staphylococcus species. Methicillin-resistant Staphylococcus pseudintermedius (MRSP) in animals has been reported to comprise 67% of total S. pseudintermedius infections and in humans it constitutes a prominent risk of drug resistant zoonotic infection transmission. MRSP infections or carriage can occur due to hospitalization, frequent visits to veterinary practices, and use of antimicrobial agents. MRSP can contaminate, colonize or infect animals. Reviews have showed increased resistance in MRSP isolates [17], [91]. Resistance to other classes of antimicrobials are not unusual for Staphylococcus genera, and the same is reported for S. pseudintermedius. Fluoroquinolones, chloramphenicol, and aminoglycoside (Amikacin) are among the classes which have been reported to be ineffective against MDR isolates. 37% of MRSP clinical isolates in dogs were reported to be resistant to amikacin in the USA [92]. However, screening of drug resistance in S. pseudintermedius is an ongoing topic of research, as the isolates are not well-studied in humans. In a case series reported by Somayaji [7], 22.2% of S. pseudintermedius human-origin isolates were resistant to methicillin and other classes of antibiotics. The minimum inhibitory concentration (MIC) of almost of all antibiotic classes was much higher in the case of S. pseudintermedius biofilm as well as in static conditions, with the exception of Rifampicin, which cannot be always the choice of drug in treatment. Drug resistance in biofilm towards “last-resort” antibiotics such as Vancomycin, Linezolid, Tigecyclin is significant, comparable to biofilm produced by other species in the Staphylococcus genus. In hospital settings clinicians might not have any other range of possibilities to treat with antibiotics if this pathogen exhibits higher MIC values to Vancomycin, Linezolid, Tigecyclin than recommended doses as these are the last resort of antibiotics available [59].
Methicillin-resistant Staphylococcus pseudintermedius (MRSP) screening
Disk-diffusion and broth microdilution tests are the most commonly used phenotypic method for antimicrobial susceptibility testing. For methicillin-resistance screening in Staphylococcus species, oxacillin or cefoxitin are used as surrogate markers, because they are sensitive and more stable. In 2018, the Clinical and Laboratory Standards Institute (CLSI document M100-28), with interpretive criteria for the determination of in vitro antimicrobial susceptibility of MRSP for isolates from humans, neither the cefoxitin minimum inhibitory concentration (MIC) nor cefoxitin disk tests are reliable for detecting mecA-mediated resistance in S. pseudintermedius, as they may produce an unacceptably high percentage of false-negative results. This guideline advises that screening for methicillin resistance should be performed by using oxacillin 1 μg disk diffusion or the MIC breakpoints as neither cefoxitin MIC, nor cefoxitin disk tests are reliable for detecting mecA mediated resistance for S. pseudintermedius (i.e., resistance in the case of ≥0.5 μg/mL of oxacillin for agar and broth dilution and ≤17 mm for disk diffusion) [93]. PCR targeting the mecA gene is the most reliable test for detecting methicillin resistance, but the equipment needed is available in only a few laboratories. The PBP2a latex agglutination test is not reliable and not recommended, as it can result in false-positive results.
Treatment
The treatment of MRSP is difficult, as there are no pre-existing guidelines or data available on the drug resistance pattern. Animal isolates have shown high prevalence of drug resistance to almost all classes of antibiotics. In European studies [94], [95], animal isolates were screened for genes responsible for drug resistance among different classes of antibiotics; these isolates were found to be positive for all genes similar to those found in S. aureus. The high level of drug resistance in S. pseudintermedius limits treatment options. Three of 24 strains were diagnosed as MRSP in a study by Somayaji et al [7]. There was no specific treatment for these patients, who were managed as outpatients. The pattern showed resistance against antimicrobial classes such as macrolides, sulfonamides, and fluoroquinolones [7]. Decolonization in animals may be achieved by with products containing chlorhexidine. Since the MRSP infections exhibit a wide spectrum of clinical symptoms and manifestations, a structured, tailored treatment plan is required, taking into account severity of disease, comorbid conditions and hospitalization.
The resistance to antibiotics is directly proportional to antibiotic use. This is the point where clinicians have more control. To prevent further antibiotic resistance, the European Wound Management Association released position document which emphasize providing an optimal environment to promote rapid healing, restricting antibiotic use to situations where they are specifically indicated, and appropriate use to reduce antibiotic resistance[96]. Certain critical antibiotics for the treatment of MRSA in humans, such as mupirocin, are legally restricted to animals in some European countries [97]. 90% of dogs who underwent treatment for MRSP, also showed resistant to antibiotic classes approved for use in humans (ciprofloxacin, clindamycin, erythromycin, kanamycin, streptomycin, and trimethoprim). Rifampicin resistance was observed in 9 out of 10 S. pseudintermedius isolates [97], [92]. Frequent use of these drugs can increase the risk of developing antibiotic resistance, thus minimizing treatment options even for human cases.
Conclusions
Although S. pseudintermedius is a known colonizer in dogs, its sudden emergence in humans is cause for concern [5]. Since the literature suggests invasive infections occurred in humans, S. pseudintermedius certainly has the potential to be virulent in human hosts [5], [7], [24], [84], [88]. It is associated with implant, skin and wound infections. Correct identification with use of rapid, easy-to-use tools is required to produce a large database for future studies and establish management guidelines for infections caused by S. pseudintermedius. As data is still lacking, conclusion about its pathogenesis are not yet possible, so that all potentially pathogenic factors should be monitored before starting treatment. Both in vitro and in vivo studies are needed to establish the connection between virulence factors. From a microbiological perspective, all possible methods should be used to differentiate S. pseudintermedius from SIG. and drug resistance patterns need to be documented for future studies. More research is required to identify and establish the link between pathogenesis and clinical disease caused by S. pseudintermedius correlation in case of human. Strict indications should apply when using antibiotics to treat animals. Treatment of S. pseudintermedius in animals is very critical, as there are no guidelines yet on using human antibiotics for treatment, which might increase the drug resistance level in these strains. So far its colonization has not been reported from humans, but it is advisable to screen patients for this pathogen when coagulase-positive species of Staphylococcus are isolated, especially from wound or skin infections. There is urgent need to establish guidelines to treat animals so the emergence of drug resistance can be stopped.
Notes
Competing interests
The authors declare that they have no competing interests.
References
[1] Kloos WE, Bannerman TL. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994 Jan;7(1):117-40. DOI: 10.1128/cmr.7.1.117[2] Hajek V. Staphylococcus intermedius, a new species isolated from animals. Int J Syst Bacteriol. 1976;26:401-08. DOI: 10.1099/00207713-26-4-401
[3] Rubin JE, Chirino-Trejo M. Prevalence, sites of colonization, and antimicrobial resistance among Staphylococcus pseudintermedius isolated from healthy dogs in Saskatoon, Canada. J Vet Diagn Invest. 2011 Mar;23(2):351-4. DOI: 10.1177/104063871102300227
[4] Talan DA, Staatz D, Staatz A, Overturf GD. Frequency of Staphylococcus intermedius as human nasopharyngeal flora. J Clin Microbiol. 1989 Oct;27(10):2393. DOI: 10.1128/JCM.27.10.2393-.1989
[5] Van Hoovels L, Vankeerberghen A, Boel A, Van Vaerenbergh K, De Beenhouwer H. First case of Staphylococcus pseudintermedius infection in a human. J Clin Microbiol. 2006 Dec;44(12):4609-12. DOI: 10.1128/JCM.01308-06
[6] Savini V, Barbarini D, Polakowska K, Gherardi G, Białecka A, Kasprowicz A, Polilli E, Marrollo R, Di Bonaventura G, Fazii P, D’Antonio D, Miedzobrodzki J, Carretto E. Methicillin-resistant Staphylococcus pseudintermedius infection in a bone marrow transplant recipient. J Clin Microbiol. 2013 May;51(5):1636-8. DOI: 10.1128/JCM.03310-12
[7] Somayaji R, Priyantha MA, Rubin JE, Church D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn Microbiol Infect Dis. 2016 Aug;85(4):471-6. DOI: 10.1016/j.diagmicrobio.2016.05.008
[8] Perreten V, Kadlec K, Schwarz S, Grönlund Andersson U, Finn M, Greko C, Moodley A, Kania SA, Frank LA, Bemis DA, Franco A, Iurescia M, Battisti A, Duim B, Wagenaar JA, van Duijkeren E, Weese JS, Fitzgerald JR, Rossano A, Guardabassi L. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother. 2010 Jun;65(6):1145-54. DOI: 10.1093/jac/dkq078
[9] Ruscher C, Lübke-Becker A, Semmler T, Wleklinski CG, Paasch A, Soba A, Stamm I, Kopp P, Wieler LH, Walther B. Widespread rapid emergence of a distinct methicillin- and multidrug-resistant Staphylococcus pseudintermedius (MRSP) genetic lineage in Europe. Vet Microbiol. 2010 Aug;144(3-4):340-6. DOI: 10.1016/j.vetmic.2010.01.008
[10] Black CC, Solyman SM, Eberlein LC, Bemis DA, Woron AM, Kania SA. Identification of a predominant multilocus sequence type, pulsed-field gel electrophoresis cluster, and novel staphylococcal chromosomal cassette in clinical isolates of mecA-containing, methicillin-resistant Staphylococcus pseudintermedius. Vet Microbiol. 2009 Nov;139(3-4):333-8. DOI: 10.1016/j.vetmic.2009.06.029
[11] Fazakerley J, Williams N, Carter S, McEwan N, Nuttall T. Heterogeneity of Staphylococcus pseudintermedius isolates from atopic and healthy dogs. Vet Dermatol. 2010 Dec;21(6):578-85. DOI: 10.1111/j.1365-3164.2010.00894.x
[12] Vincze S, Paasch A, Walther B, Ruscher C, Lübke-Becker A, Wieler LH, Barbara K. Multidrug- and methicillin resistant Staphylococcus pseudintermedius as a cause of canine pyoderma: a case report. Berl Munch Tierarztl Wochenschr. 2010 Sep-Oct;123(9-10):353-8.
[13] Soedarmanto I, Kanbar T, Ülbegi-Mohyla H, Hijazin M, Alber J, Lämmler C, Akineden Ö, Weiss R, Moritz A, Zschöck M. Genetic relatedness of methicillin-resistant Staphylococcus pseudintermedius (MRSP) isolated from a dog and the dog owner. Res Vet Sci. 2011 Dec;91(3):e25-7. DOI: 10.1016/j.rvsc.2011.01.027
[14] Bannoehr J, Franco A, Iurescia M, Battisti A, Fitzgerald JR. Molecular diagnostic identification of Staphylococcus pseudintermedius. J Clin Microbiol. 2009 Feb;47(2):469-71. DOI: 10.1128/JCM.01915-08
[15] Bannoehr J, Ben Zakour NL, Waller AS, Guardabassi L, Thoday KL, van den Broek AH, Fitzgerald JR. Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J Bacteriol. 2007 Dec;189(23):8685-92. DOI: 10.1128/JB.01150-07
[16] Solyman SM, Black CC, Duim B, van Duijkeren E, Wagenaar JA, Eblerlein LC, et al. Multilocus sequence typing (MLST) for characterization of methicillin-resistant and methicillin- susceptible clones of Staphylococcus pseudintermedius, in Speaker Abstract S2:5. In: American Society for Microbiology, editor. 2nd ASM-ESCMID Conference on Methicillin-resistant Staphylococci in Animals: Veterinary and Public Health Implications. Washington, 2011 Sep 8-11. Washington, DC: ASM;17-18. Available from: https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&ved=2ahUKEwiagrbM7bXrAhWMOcAKHR9mDxUQFjACegQIBBAB&url=https%3A%2F%2Fwww.escmid.org%2Fescmid_publications%2Fescmid_elibrary%2Fmaterial%2F%3Fmid%3D11144&usg=AOvVaw3Xqxu5h_zS2aduh5zADLyB
[17] Moodley A, Stegger M, Ben Zakour NL, Fitzgerald JR, Guardabassi L. Tandem repeat sequence analysis of staphylococcal protein A (spa) gene in methicillin-resistant Staphylococcus pseudintermedius. Vet Microbiol. 2009 Mar;135(3-4):320-6. DOI: 10.1016/j.vetmic.2008.09.070
[18] Decristophoris P, Fasola A, Benagli C, Tonolla M, Petrini O. Identification of Staphylococcus intermedius Group by MALDI-TOF MS. Syst Appl Microbiol. 2011 Feb;34(1):45-51. DOI: 10.1016/j.syapm.2010.11.004
[19] Sasaki T, Kikuchi K, Tanaka Y, Takahashi N, Kamata S, Hiramatsu K. Reclassification of phenotypically identified staphylococcus intermedius strains. J Clin Microbiol. 2007 Sep;45(9):2770-8. DOI: 10.1128/JCM.00360-07
[20] Bannoehr J, Guardabassi L. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet Dermatol. 2012 Aug;23(4):253-66, e51-2. DOI: 10.1111/j.1365-3164.2012.01046.x
[21] Garbacz K, Zarnowska S, Piechowicz L, Haras K. Pathogenicity potential of Staphylococcus pseudintermedius strains isolated from canine carriers and from dogs with infection signs. Virulence. 2013 Apr;4(3):255-9. DOI: 10.4161/viru.23526
[22] Iyori K, Futagawa-Saito K, Hisatsune J, Yamamoto M, Sekiguchi M, Ide K, Son WG, Olivry T, Sugai M, Fukuyasu T, Iwasaki T, Nishifuji K. Staphylococcus pseudintermedius exfoliative toxin EXI selectively digests canine desmoglein 1 and causes subcorneal clefts in canine epidermis. Vet Dermatol. 2011 Aug;22(4):319-26. DOI: 10.1111/j.1365-3164.2011.00952.x
[23] Iyori K, Hisatsune J, Kawakami T, Shibata S, Murayama N, Ide K, Nagata M, Fukata T, Iwasaki T, Oshima K, Hattori M, Sugai M, Nishifuji K. Identification of a novel Staphylococcus pseudintermedius exfoliative toxin gene and its prevalence in isolates from canines with pyoderma and healthy dogs. FEMS Microbiol Lett. 2010 Nov;312(2):169-75. DOI: 10.1111/j.1574-6968.2010.02113.x
[24] Tanabe T, Toyoguchi M, Hirano F, Chiba M, Onuma K, Sato H. Prevalence of staphylococcal enterotoxins in Staphylococcus pseudintermedius isolates from dogs with pyoderma and healthy dogs. Microbiol Immunol. 2013 Sep;57(9):651-4. DOI: 10.1111/1348-0421.12069
[25] Bannoehr J, Brown JK, Shaw DJ, Fitzgerald RJ, van den Broek AH, Thoday KL. Staphylococccus pseudintermedius surface proteins SpsD and SpsO mediate adherence to ex vivo canine corneocytes. Vet Dermatol. 2012 Apr;23(2):119-24, e26. DOI: 10.1111/j.1365-3164.2011.01021.x
[26] Pietrocola G, Gianotti V, Richards A, Nobile G, Geoghegan JA, Rindi S, Monk IR, Bordt AS, Foster TJ, Fitzgerald JR, Speziale P. Fibronectin Binding Proteins SpsD and SpsL Both Support Invasion of Canine Epithelial Cells by Staphylococcus pseudintermedius. Infect Immun. 2015 Oct;83(10):4093-102. DOI: 10.1128/IAI.00542-15
[27] Wladyka B, Piejko M, Bzowska M, Pieta P, Krzysik M, Mazurek Ł, Guevara-Lora I, Bukowski M, Sabat AJ, Friedrich AW, Bonar E, Międzobrodzki J, Dubin A, Mak P. A peptide factor secreted by Staphylococcus pseudintermedius exhibits properties of both bacteriocins and virulence factors. Sci Rep. 2015 Sep;5:14569. DOI: 10.1038/srep14569
[28] Gharsa H, Ben Slama K, Gómez-Sanz E, Lozano C, Klibi N, Jouini A, Messadi L, Boudabous A, Torres C. Antimicrobial resistance, virulence genes, and genetic lineages of Staphylococcus pseudintermedius in healthy dogs in tunisia. Microb Ecol. 2013 Aug;66(2):363-8. DOI: 10.1007/s00248-013-0243-y
[29] Futagawa-Saito K, Ba-Thein W, Sakurai N, Fukuyasu T. Prevalence of virulence factors in Staphylococcus intermedius isolates from dogs and pigeons. BMC Vet Res. 2006 Jan;2:4. DOI: 10.1186/1746-6148-2-4
[30] Edwards VM, Deringer JR, Callantine SD, Deobald CF, Berger PH, Kapur V, Stauffacher CV, Bohach GA. Characterization of the canine type C enterotoxin produced by Staphylococcus intermedius pyoderma isolates. Infect Immun. 1997 Jun;65(6):2346-52. DOI: 10.1128/IAI.65.6.2346-2352.1997
[31] Prevost G, Bouakham T, Piemont Y, Monteil H. Characterisation of a synergohymenotropic toxin produced by Staphylococcus intermedius. FEBS Lett. 1995 Dec;376(3):135-40. DOI: 10.1016/0014-5793(95)01260-9
[32] Couto N, Belas A, Oliveira M, Almeida P, Clemente C, Pomba C. Comparative RNA-seq-Based Transcriptome Analysis of the Virulence Characteristics of Methicillin-Resistant and -Susceptible Staphylococcus pseudintermedius Strains Isolated from Small Animals. Antimicrob Agents Chemother. 2016 Feb;60(2):962-7. DOI: 10.1128/AAC.01907-15
[33] Dziewanowska K, Edwards VM, Deringer JR, Bohach GA, Guerra DJ. Comparison of the beta-toxins from Staphylococcus aureus and Staphylococcus intermedius. Arch Biochem Biophys. 1996 Nov;335(1):102-8. DOI: 10.1006/abbi.1996.0486
[34] Futagawa-Saito K, Sugiyama T, Karube S, Sakurai N, Ba-Thein W, Fukuyasu T. Prevalence and characterization of leukotoxin-producing Staphylococcus intermedius in Isolates from dogs and pigeons. J Clin Microbiol. 2004 Nov;42(11):5324-6. DOI: 10.1128/JCM.42.11.5324-5326.2004
[35] Ruscher C, Lübke-Becker A, Wleklinski CG, Soba A, Wieler LH, Walther B. Prevalence of Methicillin-resistant Staphylococcus pseudintermedius isolated from clinical samples of companion animals and equidaes. Vet Microbiol. 2009 Apr;136(1-2):197-201. DOI: 10.1016/j.vetmic.2008.10.023
[36] Terauchi R, Sato H, Hasegawa T, Yamaguchi T, Aizawa C, Maehara N. Isolation of exfoliative toxin from Staphylococcus intermedius and its local toxicity in dogs. Vet Microbiol. 2003 Jun;94(1):19-29. DOI: 10.1016/s0378-1135(03)00048-8
[37] Terauchi R, Sato H, Endo Y, Aizawa C, Maehara N. Cloning of the gene coding for Staphylococcus intermedius exfoliative toxin and its expression in Escherichia coli. Vet Microbiol. 2003 Jun;94(1):31-8. DOI: 10.1016/s0378-1135(03)00047-6
[38] Lautz S, Kanbar T, Alber J, Lämmler C, Weiss R, Prenger-Berninghoff E, Zschöck M. Dissemination of the gene encoding exfoliative toxin of Staphylococcus intermedius among strains isolated from dogs during routine microbiological diagnostics. J Vet Med B Infect Dis Vet Public Health. 2006 Nov;53(9):434-8. DOI: 10.1111/j.1439-0450.2006.00999.x
[39] Yoon JW, Lee GJ, Lee SY, Park C, Yoo JH, Park HM. Prevalence of genes for enterotoxins, toxic shock syndrome toxin 1 and exfoliative toxin among clinical isolates of Staphylococcus pseudintermedius from canine origin. Vet Dermatol. 2010 Oct;21(5):484-9. DOI: 10.1111/j.1365-3164.2009.00874.x
[40] Futagawa-Saito K, Makino S, Sunaga F, Kato Y, Sakurai-Komada N, Ba-Thein W, Fukuyasu T. Identification of first exfoliative toxin in Staphylococcus pseudintermedius. FEMS Microbiol Lett. 2009 Dec;301(2):176-80. DOI: 10.1111/j.1574-6968.2009.01823.x
[41] Futagawa-Saito K, Suzuki M, Ohsawa M, Ohshima S, Sakurai N, Ba-Thein W, Fukuyasu T. Identification and prevalence of an enterotoxin-related gene, se-int, in Staphylococcus intermedius isolates from dogs and pigeons. J Appl Microbiol. 2004;96(6):1361-6. DOI: 10.1111/j.1365-2672.2004.02264.x
[42] Hendricks A, Schuberth HJ, Schueler K, Lloyd DH. Frequency of superantigen-producing Staphylococcus intermedius isolates from canine pyoderma and proliferation-inducing potential of superantigens in dogs. Res Vet Sci. 2002 Dec;73(3):273-7. DOI: 10.1016/s0034-5288(02)00107-8
[43] Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541-64. DOI: 10.1146/annurev.genet.42.110807.091640
[44] Dufour P, Jarraud S, Vandenesch F, Greenland T, Novick RP, Bes M, Etienne J, Lina G. High genetic variability of the agr locus in Staphylococcus species. J Bacteriol. 2002 Feb;184(4):1180-6. DOI: 10.1128/jb.184.4.1180-1186.2002
[45] Ben Zakour NL, Bannoehr J, van den Broek AH, Thoday KL, Fitzgerald JR. Complete genome sequence of the canine pathogen Staphylococcus pseudintermedius. J Bacteriol. 2011 May;193(9):2363-4. DOI: 10.1128/JB.00137-11
[46] Tse H, Tsoi HW, Leung SP, Urquhart IJ, Lau SK, Woo PC, Yuen KY. Complete genome sequence of the veterinary pathogen Staphylococcus pseudintermedius strain HKU10-03, isolated in a case of canine pyoderma. J Bacteriol. 2011 Apr;193(7):1783-4. DOI: 10.1128/JB.00023-11
[47] Simou C, Hill PB, Forsythe PJ, Thoday KL. Species specificity in the adherence of staphylococci to canine and human corneocytes: a preliminary study. Vet Dermatol. 2005 Jun;16(3):156-61. DOI: 10.1111/j.1365-3164.2005.00452.x
[48] Woolley KL, Kelly RF, Fazakerley J, Williams NJ, Nuttall TJ, McEwan NA. Reduced in vitro adherence of Staphylococcus species to feline corneocytes compared to canine and human corneocytes. Vet Dermatol. 2008 Feb;19(1):1-6. DOI: 10.1111/j.1365-3164.2007.00649.x
[49] Lu YF, McEwan NA. Staphylococcal and micrococcal adherence to canine and feline corneocytes: quantification using a simple adhesion assay. Vet Dermatol. 2007 Feb;18(1):29-35. DOI: 10.1111/j.1365-3164.2007.00567.x
[50] Forsythe PJ, Hill PB, Thoday KL, Brown J. Use of computerized image analysis to quantify staphylococcal adhesion to canine corneocytes: does breed and body site have any relevance to the pathogenesis of pyoderma? Vet Dermatol. 2002 Feb;13(1):29-36. DOI: 10.1046/j.0959-4493.2001.00269.x
[51] Saijonmaa-Koulumies LE, Lloyd DH. Adherence of Staphylococcus intermedius to canine corneocytes in vitro. Vet Dermatol. 2002 Aug;13(4):169-76. DOI: 10.1046/j.1365-3164.2002.00294.x
[52] Ben Zakour NL, Guinane CM, Fitzgerald JR. Pathogenomics of the staphylococci: insights into niche adaptation and the emergence of new virulent strains. FEMS Microbiol Lett. 2008 Dec;289(1):1-12. DOI: 10.1111/j.1574-6968.2008.01384.x
[53] Bannoehr J, Ben Zakour NL, Reglinski M, Inglis NF, Prabhakaran S, Fossum E, Smith DG, Wilson GJ, Cartwright RA, Haas J, Hook M, van den Broek AH, Thoday KL, Fitzgerald JR. Genomic and surface proteomic analysis of the canine pathogen Staphylococcus pseudintermedius reveals proteins that mediate adherence to the extracellular matrix. Infect Immun. 2011 Aug;79(8):3074-86. DOI: 10.1128/IAI.00137-11
[54] Foster TJ, Höök M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998 Dec;6(12):484-8. DOI: 10.1016/s0966-842x(98)01400-0
[55] Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SV. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003 Oct;115(2):217-28. DOI: 10.1016/s0092-8674(03)00809-2
[56] Cree RG, Noble WC. In vitro indices of tissue adherence in Staphylococcus intermedius. Lett Appl Microbiol. 1995 Mar;20(3):168-70. DOI: 10.1111/j.1472-765x.1995.tb00418.x
[57] Geoghegan JA, Smith EJ, Speziale P, Foster TJ. Staphylococcus pseudintermedius expresses surface proteins that closely resemble those from Staphylococcus aureus. Vet Microbiol. 2009 Sep;138(3-4):345-52. DOI: 10.1016/j.vetmic.2009.03.030
[58] Gangil R, Audarya S, Chhabra D, Sikrodia R, Sharda R. Multi Drug Resistant Staphylococcus pseudintermedius isolates from Canine Pyoderma. Ind J Anim Res. 2015;5(4):749-52. DOI: 10.5958/2277-940X.2015.00124.2
[59] Pompilio A, De Nicola S, Crocetta V, Guarnieri S, Savini V, Carretto E, Di Bonaventura G. New insights in Staphylococcus pseudintermedius pathogenicity: antibiotic-resistant biofilm formation by a human wound-associated strain. BMC Microbiol. 2015 May;15:109. DOI: 10.1186/s12866-015-0449-x
[60] Duim B, Verstappen KMHW, Kalupahana RS, Ranathunga L, Fluit AC, Wagenaar JA. Methicillin-resistant Staphylococcus pseudintermedius among dogs in the description of novel SCCmec variants. Vet Microbiol. 2018 Jan;213:136-41. DOI: 10.1016/j.vetmic.2017.11.022
[61] Phumthanakorn N, Fungwithaya P, Chanchaithong P, Prapasarakul N. Enterotoxin gene profile of methicillin-resistant Staphylococcus pseudintermedius isolates from dogs, humans and the environment. J Med Microbiol. 2018 Jun;67(6):866-73. DOI: 10.1099/jmm.0.000748
[62] Feng Y, Tian W, Lin D, Luo Q, Zhou Y, Yang T, Deng Y, Liu YH, Liu JH. Prevalence and characterization of methicillin-resistant Staphylococcus pseudintermedius in pets from South China. Vet Microbiol. 2012 Dec;160(3-4):517-24. DOI: 10.1016/j.vetmic.2012.06.015
[63] Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, Hirotaki S, Kawakami T, Fukata T, Hiramatsu K. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol. 2010 Mar;48(3):765-9. DOI: 10.1128/JCM.01232-09
[64] Kang JH, Chung TH, Hwang CY. Clonal distribution of methicillin-resistant Staphylococcus pseudintermedius isolates from skin infection of dogs in Korea. Vet Microbiol. 2017 Oct;210:32-7. DOI: 10.1016/j.vetmic.2017.08.017
[65] Han JI, Rhim H, Yang CH, Park HM. Molecular characteristics of new clonal complexes of Staphylococcus pseudintermedius from clinically normal dogs. Vet Q. 2018 Dec;38(1):14-20. DOI: 10.1080/01652176.2017.1400710
[66] Worthing KA, Marcus A, Abraham S, Trott DJ, Norris JM. Qac genes and biocide tolerance in clinical veterinary methicillin-resistant and methicillin-susceptible Staphylococcus aureus and Staphylococcus pseudintermedius. Vet Microbiol. 2018 Mar;216:153-8. DOI: 10.1016/j.vetmic.2018.02.004
[67] Youn JH, Park YH, Hang'ombe B, Sugimoto C. Prevalence and characterization of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from companion animals and environment in the veterinary teaching hospital in Zambia, Africa. Comp Immunol Microbiol Infect Dis. 2014 Mar;37(2):123-30. DOI: 10.1016/j.cimid.2014.01.003
[68] Summers JF, Hendricks A, Brodbelt DC. Prescribing practices of primary-care veterinary practitioners in dogs diagnosed with bacterial pyoderma. BMC Vet Res. 2014 Oct;10:240. DOI: 10.1186/s12917-014-0240-5
[69] McCarthy AJ, Harrison EM, Stanczak-Mrozek K, Leggett B, Waller A, Holmes MA, Lloyd DH, Lindsay JA, Loeffler A. Genomic insights into the rapid emergence and evolution of MDR in Staphylococcus pseudintermedius. J Antimicrob Chemother. 2015 Apr;70(4):997-1007. DOI: 10.1093/jac/dku496
[70] Kjellman EE, Slettemeås JS, Small H, Sunde M. Methicillin-resistant Staphylococcus pseudintermedius (MRSP) from healthy dogs in Norway - occurrence, genotypes and comparison to clinical MRSP. Microbiologyopen. 2015 Dec;4(6):857-66. DOI: 10.1002/mbo3.258
[71] Worthing KA, Abraham S, Coombs GW, Pang S, Saputra S, Jordan D, Trott DJ, Norris JM. Clonal diversity and geographic distribution of methicillin-resistant Staphylococcus pseudintermedius from Australian animals: Discovery of novel sequence types. Vet Microbiol. 2018 Jan;213:58-65. DOI: 10.1016/j.vetmic.2017.11.018
[72] Grönthal T, Eklund M, Thomson K, Piiparinen H, Sironen T, Rantala M. Antimicrobial resistance in Staphylococcus pseudintermedius and the molecular epidemiology of methicillin-resistant S. pseudintermedius in small animals in Finland. J Antimicrob Chemother. 2017 07;72(7):2141. DOI: 10.1093/jac/dkx086
[73] Verstappen KM, Huijbregts L, Spaninks M, Wagenaar JA, Fluit AC, Duim B. Development of a real-time PCR for detection of Staphylococcus pseudintermedius using a novel automated comparison of whole-genome sequences. PLoS ONE. 2017;12(8):e0183925. DOI: 10.1371/journal.pone.0183925
[74] Kizerwetter-Świda M, Chrobak-Chmiel D, Rzewuska M, Binek M. Changes in the population structure of canine methicillin-resistant Staphylococcus pseudintermedius in Poland. Vet Microbiol. 2017 Sep;208:106-9. DOI: 10.1016/j.vetmic.2017.07.025
[75] Kmieciak W, Szewczyk EM, Ciszewski M. Searching for Beta-Haemolysin hlb Gene in Staphylococcus pseudintermedius with Species-Specific Primers. Curr Microbiol. 2016 Jul;73(1):148-52. DOI: 10.1007/s00284-016-1038-4
[76] Bannoehr J, Guardabassi L. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet Dermatol. 2012 Aug;23(4):253-66, e51-2. DOI: 10.1111/j.1365-3164.2012.01046.x
[77] Pires Dos Santos T, Damborg P, Moodley A, Guardabassi L. Systematic Review on Global Epidemiology of Methicillin-Resistant: Inference of Population Structure from Multilocus Sequence Typing Data. Front Microbiol. 2016;7:1599. DOI: 10.3389/fmicb.2016.01599
[78] Börjesson S, Landén A, Bergström M, Andersson UG. Methicillin-resistant Staphylococcus pseudintermedius in Sweden. Microb Drug Resist. 2012 Dec;18(6):597-603. DOI: 10.1089/mdr.2012.0069
[79] Lehner G, Linek M, Bond R, Lloyd DH, Prenger-Berninghoff E, Thom N, Straube I, Verheyen K, Loeffler A. Case-control risk factor study of methicillin-resistant Staphylococcus pseudintermedius (MRSP) infection in dogs and cats in Germany. Vet Microbiol. 2014 Jan;168(1):154-60. DOI: 10.1016/j.vetmic.2013.10.023
[80] Haenni M, de Moraes NA, Châtre P, Médaille C, Moodley A, Madec JY. Characterisation of clinical canine meticillin-resistant and meticillin-susceptible Staphylococcus pseudintermedius in France. J Glob Antimicrob Resist. 2014 Jun;2(2):119-23. DOI: 10.1016/j.jgar.2014.02.002
[81] Ventrella G, Moodley A, Grandolfo E, Parisi A, Corrente M, Buonavoglia D, Guardabassi L. Frequency, antimicrobial susceptibility and clonal distribution of methicillin-resistant Staphylococcus pseudintermedius in canine clinical samples submitted to a veterinary diagnostic laboratory in Italy: A 3-year retrospective investigation. Vet Microbiol. 2017 Nov;211:103-6. DOI: 10.1016/j.vetmic.2017.09.015
[82] Videla R, Solyman SM, Brahmbhatt A, Sadeghi L, Bemis DA, Kania SA. Clonal Complexes and Antimicrobial Susceptibility Profiles of Staphylococcus pseudintermedius Isolates from Dogs in the United States. Microb Drug Resist. 2018 Jan/Feb;24(1):83-8. DOI: 10.1089/mdr.2016.0250
[83] Riegel P, Jesel-Morel L, Laventie B, Boisset S, Vandenesch F, Prévost G. Coagulase-positive Staphylococcus pseudintermedius from animals causing human endocarditis. Int J Med Microbiol. 2011 Mar;301(3):237-9. DOI: 10.1016/j.ijmm.2010.09.001
[84] De Martino L, Nocera FP, Mallardo K, Nizza S, Masturzo E, Fiorito F, Iovane G, Catalanotti P. An update on microbiological causes of canine otitis externa in Campania Region, Italy. Asian Pac J Trop Biomed. 2016;6(5):384-9. DOI: 10.1016/j.apjtb.2015.11.012
[85] Maaland M, Guardabassi L. In vitro antimicrobial activity of nitrofurantoin against Escherichia coli and Staphylococcus pseudintermedius isolated from dogs and cats. Vet Microbiol. 2011 Aug;151(3-4):396-9. DOI: 10.1016/j.vetmic.2011.03.009
[86] Stegmann R, Burnens A, Maranta CA, Perreten V. Human infection associated with methicillin-resistant Staphylococcus pseudintermedius ST71. J Antimicrob Chemother. 2010 Sep;65(9):2047-8. DOI: 10.1093/jac/dkq241
[87] Weese JS, van Duijkeren E. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet Microbiol. 2010 Jan;140(3-4):418-29. DOI: 10.1016/j.vetmic.2009.01.039
[88] Bean DC, Wigmore SM. Carriage rate and antibiotic susceptibility of coagulase-positive staphylococci isolated from healthy dogs in Victoria, Australia. Aust Vet J. 2016 Dec;94(12):456-60. DOI: 10.1111/avj.12528
[89] Kang MH, Chae MJ, Yoon JW, Kim SG, Lee SY, Yoo JH, Park HM. Antibiotic resistance and molecular characterization of ophthalmic Staphylococcus pseudintermedius isolates from dogs. J Vet Sci. 2014;15(3):409-15. DOI: 10.4142/jvs.2014.15.3.409
[90] Yoon JW, Lee KJ, Lee SY, Chae MJ, Park JK, Yoo JH, Park HM. Antibiotic resistance profiles of Staphylococcus pseudintermedius isolates from canine patients in Korea. J Microbiol Biotechnol. 2010 Dec;20(12):1764-8.
[91] Grönthal T, Moodley A, Nykäsenoja S, Junnila J, Guardabassi L, Thomson K, Rantala M. Large outbreak caused by methicillin resistant Staphylococcus pseudintermedius ST71 in a Finnish Veterinary Teaching Hospital-from outbreak control to outbreak prevention. PLoS ONE. 2014;9(10):e110084. DOI: 10.1371/journal.pone.0110084
[92] Gold RM, Cohen ND, Lawhon SD. Amikacin resistance in Staphylococcus pseudintermedius isolated from dogs. J Clin Microbiol. 2014 Oct;52(10):3641-6. DOI: 10.1128/JCM.01253-14
[93] CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
[94] Grönthal T, Eklund M, Thomson K, Piiparinen H, Sironen T, Rantala M. Antimicrobial resistance in Staphylococcus pseudintermedius and the molecular epidemiology of methicillin-resistant S. pseudintermedius in small animals in Finland. J Antimicrob Chemother. 2017 04;72(4):1021-30. DOI: 10.1093/jac/dkw559
[95] Meroni G, Soares Filipe JF, Drago L, Martino PA. Investigation on Antibiotic-Resistance, Biofilm Formation and Virulence Factors in Multi Drug Resistant and Non Multi Drug Resistant. Microorganisms. 2019 Dec;7(12). DOI: 10.3390/microorganisms7120702
[96] European Medicines Agency; Committee for Medicinal Products for Veterinary Use. Advice on implementing measures under Article 37(4) of Regulation (EU) 2019/6 on veterinary medicinal products - Criteria for the designation of antimicrobials to be reserved for treatment of certain infections in humans. EMA/CVMP/158366/2019. 2019 Oct 31. Available from: https://www.ema.europa.eu/en/documents/regulatory-procedural-guideline/advice-implementing-measures-under-article-374-regulation-eu-2019/6-veterinary-medicinal-products-criteria-designation-antimicrobials-be-reserved-treatment-certain_en.pdf
[97] Moodley A, Stegger M, Ben Zakour NL, Fitzgerald JR, Guardabassi L. Tandem repeat sequence analysis of staphylococcal protein A (spa) gene in methicillin-resistant Staphylococcus pseudintermedius. Vet Microbiol. 2009 Mar;135(3-4):320-6. DOI: 10.1016/j.vetmic.2008.09.070
[98] Börjesson S, Gómez-Sanz E, Ekström K, Torres C, Grönlund U. Staphylococcus pseudintermedius can be misdiagnosed as Staphylococcus aureus in humans with dog bite wounds. Eur J Clin Microbiol Infect Dis. 2015 Apr;34(4):839-44. DOI: 10.1007/s10096-014-2300-y
[99] Kizerwetter-Świda M, Chrobak-Chmiel D, Rzewuska M, Binek M. Changes in the population structure of canine methicillin-resistant Staphylococcus pseudintermedius in Poland. Vet Microbiol. 2017 Sep;208:106-9. DOI: 10.1016/j.vetmic.2017.07.025
[100] Börjesson S, Gómez-Sanz E, Ekström K, Torres C, Grönlund U. Staphylococcus pseudintermedius can be misdiagnosed as Staphylococcus aureus in humans with dog bite wounds. Eur J Clin Microbiol Infect Dis. 2015 Apr;34(4):839-44. DOI: 10.1007/s10096-014-2300-y
[101] Chuang CY, Yang YL, Hsueh PR, Lee PI. Catheter-related bacteremia caused by Staphylococcus pseudintermedius refractory to antibiotic-lock therapy in a hemophilic child with dog exposure. J Clin Microbiol. 2010 Apr;48(4):1497-8. DOI: 10.1128/JCM.02033-09




