[Umsetzung krankenhaushygienischer Primärpräventionsmaßnahmen verhindert die Übertragung von P. aeruginosa bei Patienten mit cystischer Fibrose im Krankenhaus]
Benedikt Matt 1Dieter Mitteregger 2
Sabine Renner 3
Elisabeth Presterl 1
Ojan Assadian 1
Magda Diab-Elschahawi 1
1 Department for Hospital Hygiene and Infecion Control, Medical University of Vienna, Vienna General Hospital, Wien, Austria
2 Clinical Department for Clinical Microbiology, Medical University of Vienna, Vienna General Hospital, Wien, Austria
3 Department of Paediatrics and Adolescent Medicine, Medical University of Vienna, Vienna General Hospital, Wien, Austria
Zusammenfassung
Hintergrund: Ziel dieser Studie war es, die Übertragungsepidemiologie von Pseudomonas aeruginosa-Isolaten von stationärer Patienten mit cystischer Fibrose (CF) am Allgemeinen Krankenhaus der Stadt Wien (AKH Wien) anhand ihres molekularbiologischen Fingerabdrucks zu untersuchen, um die Mechanismen der Übertragung zwischen CF-Patienten zu identifizieren und gezielte infektionspräventive Maßnahmen zu implementieren.
Methoden: Am AKH Wien werden Kinder und Jugendliche mit CF bis zum vollendeten 18. Lebensalter betreut. Zum Studienzeitpunkt befanden sich 139 Patienten in Behandlung, von denen bei 41 Patienten P. aeruginosa im Sputum und/oder Rachenabstrich nachweisbar war. Insgesamt wurden 50 P. aeruginosa-Isolate von den 41 P. aeruginosa-positiven CF-Patienten, die zwischen August 2010 und März 2012 im Rahmen von Routineuntersuchungen isoliert worden waren, hinsichtlich ihrer molekulargenetischen Eigenschaften mittels DiversiLab® untersucht.
Ergebnisse: Es konnten 42 nicht weiter unterscheidbare molekular-biologische Muster identifiziert werden, wobei sich 7 Muster häufiger als die anderen fanden. 40 der insgesamt 42 Genotypen wurden nur von einzelnen Patienten isoliert, 2 Genotypen waren jeweils bei 2 unterschiedlichen Patienten nachweisbar. Neun Patienten wiesen 2 oder mehr phänotypisch unterschiedlicher Isolate aus, bei 5 dieser Patienten gehörten die Isolate zum selben Genotyp.
Schlussfolgerung: Die breite genetische Heterogenität der P. aeruginosa-Isolate in den untersuchten CF-Patienten weist darauf hin, dass die Mehrzahl an CF-Patienten in stationärer Behandlung am AKH Wien diese Isolate von anderen Quellen als von zur selben Zeit in Betreuung stehenden Patienten oder von einer gemeinsamen internen Umgebungsquelle erworben haben müsste. Das legt die Schlussfolgerung nahe, dass sich die an der CF-Klinik implementierten krankenhaushygienischen Maßnahmen hinsichtlich der Verhinderung einer Übertragung von CF-Patienten erfolgreich erwiesen haben. Chronische polyklonale Infektionen bzw. Kolonisationen waren innerhalb der untersuchten CF-Patienten selten.
Schlüsselwörter
Krankenhaushygiene, repetitive sequenz-basierte PCR, cystische Fibrose, Pseudomonas aeruginosa
Background
Despite major improvements in the treatment of cystic fibrosis (CF) during the past decades, CF still limits substantially both patients’ life expectancy and quality of life. While significant survival benefit is reported for patients who remain free of infection with P. aeruginosa [1], [2], chronic or exacerbating infection with P. aeruginosa is associated with a rapid deterioration of lung function [1], [2]. The emergence of multiresistant strains complicates and limits antimicrobial therapeutic options [2], [3]. Therefore, primary prevention of colonization and infection plays a vital role in the care of CF patients [2], [4].
P. aeruginosa colonization and infection may either be community-acquired or health care associated. Multiple studies have reported on the risk factors for infection and have recommended infection control measures and behavioural changes to primary prevent the transmission of P. aeruginosa [5], [6], [7], [8], [9]. In addition, as secondary preventive measures, continuous surveillance and outbreak control is of importance to minimize the prevalence and transmission of P. aeruginosa in the hospital environment [2].
Based on the comprehensive recommendations released by the Robert Koch Institute (RKI, Germany), strict guidelines and hygiene measures have been introduced at the Vienna General Hospital (VGH) to protect CF patients from both, nosocomial and community-acquired infection. These measures were complemented with continuous infection surveillance to evaluate their performance.
The aim of this study was to characterise the epidemiology of P. aeruginosa from CF patients at the VGH and to evaluate the infection control protocols and guidelines currently established at the VGH and in the patients’ homes.
Methods
Study setting
This study was conducted at the outpatient clinic for CF patients of the department of paediatrics and adolescent medicine, Vienna General Hospital (VGH). The VGH is a 2,200 bed tertiary care medical university teaching hospital. In average, 139 CF patients aged between 0 and 21 are subject to quarterly routine examinations on which occasion sputum and/or throat swabs are obtained and tested for P. aeruginosa (and other pathogens, including Staphylococcus aureus, Burkholderia cepacia complex and fungi; not considered in this study) using routine microbiological methods for culture and identification. In this study, 50 P. aeruginosa isolates of 41 CF patients obtained between August 6th 2010 and March 9th 2012 were subject to molecular biological characterization. For 9 patients more than one isolate was included in this study, in each case these isolates derived from a single sputum or throat swab and were distinguished based on their phenotypic traits (i.e. mucoid/non mucoid phenotype). The study was approved by the Ethics Committee of the Medical University of Vienna (EC No. 424/2011). All patients or their parents gave written consent.
DNA extraction
The genomic DNA from each of the 50 isolates was extracted using the UltraClean® Microbial DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA), following the manufacturer’s instructions. The concentration and purity of the harvested DNA was measured with the NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and the concentration was adjusted to approximately 30–40 ng/µl.
Molecular characterisation by automated repetitive sequence-based PCR typing system (DiversiLab®)
All DNA samples obtained from the 50 isolates were amplified by using the DiversiLab® Pseudomonas Kit (BioMerieux, Austria GmbH), following the manufacturer’s instructions. Briefly, 60–80 ng genomic DNA, 2.5 U AmpliTaq polymerase, 2.5 µl 10× PCR buffer (Applied Biosystems, California, U.S.A.) and 2 µl primer mix were added to the rep-PCR master mix in a total volume of 25 µl per reaction. Thermal cycling parameters were as follows: initial denaturation of 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s and extension at 70°C for 90 s, with a final extension at 70°C for 3 min. The amplified DNA products were separated in a microfluidics DNA LabChip® device and detected in the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA) of the DiversiLab® system. Fingerprint patterns in the form of graphs were generated and analyzed with the web-based DiversiLab® software (V3.4; bioMérieux, Inc., Durham, NC). The results were stored as dendrogram with a similarity matrix and a virtual gel image of the fingerprint for each DNA sample (Figure 1 [Fig. 1]). Isolates with ≥95% similarity were interpreted manually on the bases of their profile overlays. Those with one or less band differences in their fingerprint patterns were considered “indistinguishable” and were assigned to the same genotype.
Figure 1: Dendrogram based on percent similarities between isolates. Roman numerals summarize isolates with the same genotype according to manual interpretation.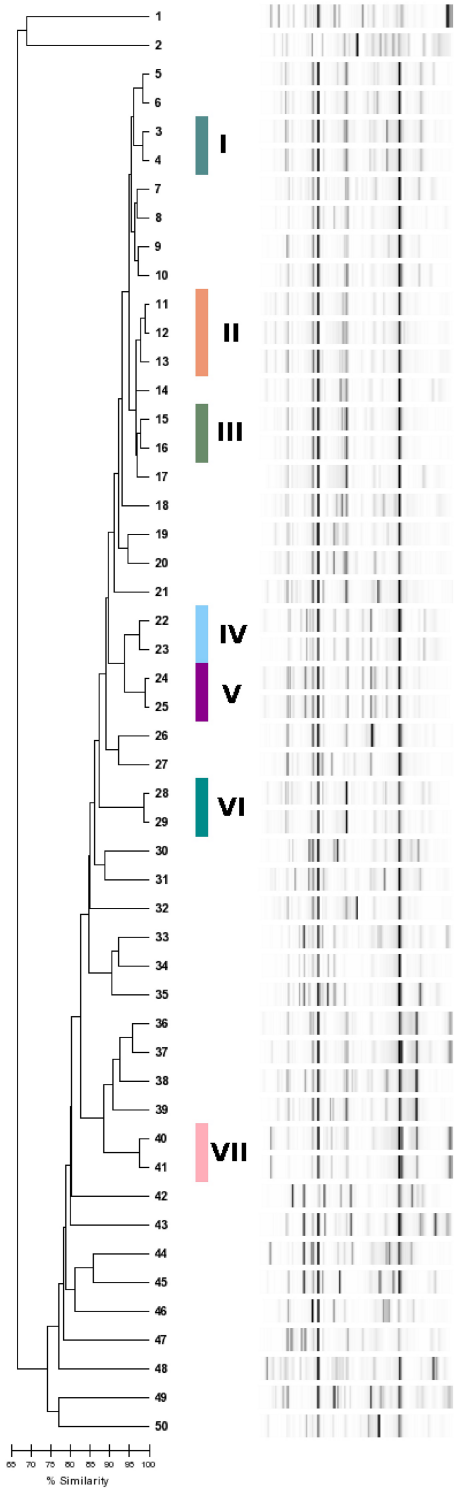
Infection control measures and guidelines for CF patients at the VGH. The following infection control measures were implemented before onset of the study: CF patients were required to wear masks on entering the hospital as well as on the ward when they were outside of their assigned rooms. It was prohibited for CF patients to visit the rooms of other CF patients. In the pulmonary function laboratory and during physiotherapy the medical personnel wear gowns and masks (and disposable gloves and plastic aprons during activities with risk of contamination). Contamination of the patients’ clothing with the patients’ own respiratory secretion was avoided. After every examination, surfaces close to the patients were disinfected.
In outpatient care CF patients with multiresistant pathogens were examined on unscheduled appointments, they entered the facility through a separate door and were assigned their own examination room. The medical personnel wear masks, gowns, disposable gloves and plastic aprons. All objects of use and surfaces were disinfected in case of contamination, the provided stethoscope was not used anywhere else. When leaving the examination room, the patients wear masks until they had left the hospital. Inpatient CF cases with multiresistant bacteria were cared for in single rooms only. Alcohol-based hand rubs, disposable gloves, gowns, masks and appropriate disposal containers were provided in the patient’s room anteroom. If the patients had to leave their rooms, hygienic hand disinfection, protection gowns and masks were compulsory. The same (except for masks) applied to relatives that have been admitted as companions. On the day of release, the patients were asked to leave the ward directly, in their street clothing and with a fresh face mask.
At home, patients were advised never to use someone else’s nebulizer or share their own nebulizer with anyone else. The contact between CF patients outside the hospital was allowed, however, limiting close contact such as sleeping in the same room or shaking hands. For patients with a temporarily high bacterial count and for those who tested positive for multiresistant pathogens, any personal contact to other CF patients was strongly discouraged.
Statistical analysis
For calculation of the percent genetic similarities between samples, the Pearson’s correlation method was used.
Results
One-hundred thirty nine patients were included in this study, of which 40 were identified with P. aeruginosa. Of those positive: 19 were male (47.5%) and 21 female (52.5%); their age ranged between 2.8 years – 21.5 years with a mean age of 11.9 years. One patient harboured three phenotypically different isolates, eight patients harboured two isolates each and 31 patients were represented by one isolate each.
Among a total of 50 typed isolates 42 distinguishable genotypes were identified, of which 35 were only observed once. Seven genotypes were found multiple times and were numbered I–VII. One genotype (II) comprised three strains that were retrieved from two different patients. Six genotypes comprised two strains each (I, III–VII). One of these (IV) comprised two isolates retrieved from two different patients while the others (I, III, V–VII) each comprised two isolates that were retrieved from single patients (Figure 1 [Fig. 1], Table 1 [Tab. 1]).
Table 1: Selection of strains with known or alleged connection and their assigned genotypes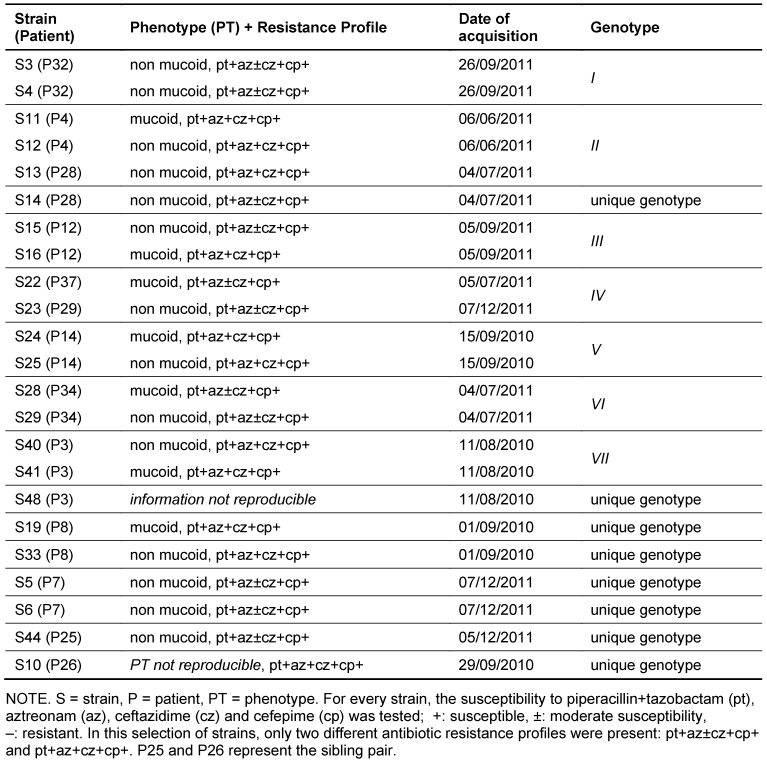
Of the eight patients with two isolates each, five harboured two isolates of the same genotype. One of the two isolates of patient 28 was assigned to genotype II together with two strains of another patient (patient 4). The other was assigned to a unique genotype. Two of the three isolates of patient 3 shared a genotype (VII). The genotype of the remaining isolate was unique (Figure 1 [Fig. 1], Table 1 [Tab. 1]).
Similarities between strains from single patients thus accounted for all but two genotypes, namely genotypes II and IV, which were shared by two different patients each.
The patient population included one sibling pair, whose strains did not belong to the same genotype.
All strains with any known/alleged epidemiological connection or genetic relationship are depicted in Table 1 [Tab. 1].
Discussion
We identified a considerable genetic heterogeneity of P. aeruginosa in our patient population. Almost every genotype was harboured by only a single patient, with the exception of genotypes II and IV, which were each present in two patients. In addition, most of our patients with alleged polyclonal infection/colonization with P. aeruginosa harboured only a single genotype exhibiting phenotypic variations.
In their review on the approach to P. aeruginosa in cystic fibrosis, Bendiak et al. [5] stated that environmental reservoirs play a major role in the acquisition of P. aeruginosa. This aspect is supported by the great genetic heterogeneity of P. aeruginosa in our patient population, which suggests that the majority of CF patients under care at the VGH acquire P. aeruginosa endogenously or from environmental sources. Our data implicates that the implemented guidelines and hygiene measures have succeeded to primary prevent nosocomial transmission of P. aeruginosa to the CF patients under care at the VGH and patient-to-patient transmission outside the hospital. These findings justify the additional workload for hospital personnel due to enhanced infection control measures, the financial burden to the health care system and the substantial psychological burden to the patients.
Transmission could be excluded for all but four patients, who shared two genotypes of P. aeruginosa, namely genotypes II (shared by patients 4 and 28) and IV (shared by patients 37 and 29, Table 1 [Tab. 1]). Unfortunately, there is limited information available regarding possible relationships or contact outside the hospital between the patients sharing these genotypes. Possible explanations range from shared activities outside the hospital to a breach in the hospital hygiene measures. However, inpatient and outpatient stays at the VGH were retraced for both patients 4 and 28, and patients 37 and 29, respectively, until a year before the isolates in question were detected in the routine examinations, and no overlapping stays were found. This makes transmissions in the hospital environment improbable, at least within this time frame.
Our findings are in concordance with a similar study published recently [10]. Masoud-Landgraf et al. typed 86 P. aeruginosa isolates from 43 CF patients under care in the CF centre of the Medical University of Graz, Austria, and found that the majority of CF patients under their investigation harboured unique P. aeruginosa genotypes. Putting our data together with results published by Masoud-Landgraf et al. and [10], it may be argued, that for the majority of CF patients in Austria nosocomial acquisition of P. aeruginosa does not constitute a relevant source of infection.
Similarly to Masoud-Landgraf et al. [10], most of our patients with alleged polyclonal infection with P. aeruginosa seemed to harbour merely two members of a single genotype exhibiting phenotypic differences, namely in their morphotypes or their antibiotic resistance profiles, possibly acquired by mutation during infection or culturing. This suggests that chronic infection with more than one genotype of P. aeruginosa is rare, at least in the population under investigation.
For genotyping the P. aeruginosa isolates under examination, the DiversiLab® method was employed. There are conflicting studies both relating to the informative value of the DiversiLab® method when used for typing of P. aeruginosa and to the cut-off value for characterisation of isolates as “linked” or “different” [11], [12], [13], [14]. According to Deplano et al. [14], each user needs to validate the DiversiLab® protocol for each species and set its type similarity cut-off values prior to routine use.
In our study, as similarly perceived by Doleans-Jordheim et al. [11], the DiversiLab® method proved to be rapid and easy to perform. It involved limited human resources. The DiversiLab® method has also shown good interpersonal reproducibility. Furthermore, with the employed criteria, the analysis produced excellent epidemiological concordance.
Limitations of our study comprise the limited availability of information on social contact such as personal friendships, shared activities and other contacts/relationships between patients. This reduced, to some degree, our ability to interpret the acquired results. Although great care was taken to minimize the time differences between the dates of acquisition of each isolate, the date of strain acquisition differs between patients by an average of 192 days (433 days in the example of the sibling pair). This might have caused us to miss transient cross-infections and epidemics. Second, we did not screen for stool carriage of P. aeruginosa within the patient population. Therefore, it cannot be ruled out that isolated P. aeruginosa strains colonized patients endogenously, and not through exogenous environmental sources.
A particular strength of this study, however, is its representativeness. All patients under care at the VGH got regularly tested for respiratory infection with P. aeruginosa. All of those who were tested positive at some point within almost two years could be included in this study except one, where not enough DNA could be harvested from the respective isolates for PCR and further analysis. Thus the acquired and studied patient population represents the population under our investigation well.
Conclusion
Our data suggest that there has been no transmission of P. aeruginosa isolates between CF patients under care at the VGH, indicating the success of the hygiene measures employed at the CF centre and the guidelines regarding contact between patients outside the hospital. The clear majority of chronic infections with a single clone of P. aeruginosa indicate that polyclonal chronic infection with P. aeruginosa is rare, at least in the population under investigation. In our study setting, the DiversiLab® method proved to be a rapid and easy-to-perform tool for typing P. aeruginosa.
We recommend that currently established infection control measures in the hospital setting as well as in CF patient homes are continuously enforced and that more research efforts be directed towards the prevention of community-acquired P. aeruginosa infections, such as the development of an effective vaccine against P. aeruginosa. An effective vaccine would considerably decrease the danger that P. aeruginosa poses to this vulnerable patient population.
Notes
Competing interests
The authors declare that they have no competing interests.
References
[1] O'Sullivan BP, Freedman SD. Cystic fibrosis. Lancet. 2009 May 30;373(9678):1891-904. DOI: 10.1016/S0140-6736(09)60327-5[2] Mandell GL, Bennett JE, Dolin R, editors. Mandell, Douglas, and Bennett's principles and practice of infectious diseases. 7th ed. Philadelphia, PA: Churchill Livingstone/Elsevier; 2010.
[3] European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2011. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). Stockholm: ECDC; 2012. DOI: 10.2900/6551
[4] Cystic Fibrosis Foundation. Patient Registry Annual Data Report. Bethesda, Maryland: Cystic Fibrosis Foundation; 2011.
[5] Bendiak GN, Ratjen F. The approach to Pseudomonas aeruginosa in cystic fibrosis. Semin Respir Crit Care Med. 2009 Oct;30(5):587-95. DOI: 10.1055/s-0029-1238917
[6] Exner M, Kramer A, Lajoie L, Gebel J, Engelhart S, Hartemann P. Prevention and control of health care-associated waterborne infections in health care facilities. Am J Infect Control. 2005 Jun;33(5 Suppl 1):S26-40. DOI: 10.1016/j.ajic.2005.04.002
[7] Festini F, Buzzetti R, Bassi C, Braggion C, Salvatore D, Taccetti G, Mastella G. Isolation measures for prevention of infection with respiratory pathogens in cystic fibrosis: a systematic review. J Hosp Infect. 2006 Sep;64(1):1-6. DOI: 10.1016/j.jhin.2006.02.021
[8] Döring G. Prevention of Pseudomonas aeruginosa infection in cystic fibrosis patients. Int J Med Microbiol. 2010 Dec;300(8):573-7. DOI: 10.1016/j.ijmm.2010.08.010
[9] Schelstraete P, Van Daele S, De Boeck K, Proesmans M, Lebecque P, Leclercq-Foucart J, Malfroot A, Vaneechoutte M, De Baets F. Pseudomonas aeruginosa in the home environment of newly infected cystic fibrosis patients. Eur Respir J. 2008 Apr;31(4):822-9. DOI: 10.1183/09031936.00088907
[10] Masoud-Landgraf L, Badura A, Eber E, Feierl G, Posch J, Zarfel G, Zach M, Marth E. Molecular epidemiology of Pseudomonas aeruginosa in cystic fibrosis patients from Southeast Austria. Wien Klin Wochenschr. 2012 Apr;124(7-8):262-5. DOI: 10.1007/s00508-012-0156-7
[11] Doléans-Jordheim A, Cournoyer B, Bergeron E, Croizé J, Salord H, André J, Mazoyer MA, Renaud FN, Freney J. Reliability of Pseudomonas aeruginosa semi-automated rep-PCR genotyping in various epidemiological situations. Eur J Clin Microbiol Infect Dis. 2009;28(9):1105-11. DOI: 10.1007/s10096-009-0755-z
[12] Ratkai C, Peixe LV, Grosso F, Freitas AR, Antunes P, Fodor E, Hajdú E, Nagy E. Successful application of the DiversiLab repetitive-sequence-based PCR typing system for confirmation of the circulation of a multiresistant Pseudomonas aeruginosa clone in different hospital wards. Diagn Microbiol Infect Dis. 2010 Jun;67(2):202-6. DOI: 10.1016/j.diagmicrobio.2010.01.010
[13] Fluit AC, Terlingen AM, Andriessen L, Ikawaty R, van Mansfeld R, Top J, Cohen Stuart JW, Leverstein-van Hall MA, Boel CH. Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species. J Clin Microbiol. 2010 Nov;48(11):3979-89. DOI: 10.1128/JCM.01191-10
[14] Deplano A, Denis O, Rodriguez-Villalobos H, De Ryck R, Struelens MJ, Hallin M. Controlled performance evaluation of the DiversiLab repetitive-sequence-based genotyping system for typing multidrug-resistant health care-associated bacterial pathogens. J Clin Microbiol. 2011 Oct;49(10):3616-20. DOI: 10.1128/JCM.00528-11




