[Prävalenz und klonale Abstammungslinien von Biofilm produzierenden Staphylococcus aureus Stämmen aus klinischen Proben und von medizinischem Personal im Ahmadu Bello University Teaching Hospital, Nigeria]
Kabir Umar 1Idris Nasir Abdullahi 1,2
Abdulkadir Magaji Magashi 3
Abdullahi Hassan Kawo 3
Yahaya Usman 1
Abdurrahaman El-fulaty Ahmad 1
Carmen Torres 2
1 Department of Medical Laboratory Science, Faculty of Allied Health Sciences, Ahmadu Bello University, Zaria, Nigeria
2 Area of Biochemistry and Molecular Biology, OneHealth-UR Research Group, University of La Rioja, Logroño, Spain
3 Department of Microbiology, Faculty of Life Sciences, Bayero University, Kano, Nigeria
Zusammenfassung
In der Studie wurden die Häufigkeit und die molekularen Merkmale von Staphylococcus (S.) aureus bei 206 Patienten mit Verbrennungen und Wunden sowie bei 94 Mitarbeitern des Ahmadu Bello University Teaching Hospital, Zaria, Nordnigeria, analysiert. In neun (4,4%) Proben von Verbrennungspatienten und fünf (5,3%) von medizinischem Personal wurde S. aureus nachgewiesen. Sieben (50%) waren mecA-positiv (assoziiert mit SCCmec-Typen IVa und V), 35,7% wiesen einen multiresistenten (MDR) Phänotyp auf. Die S. aureus Isolate gehörten zu 11 verschiedenen Spa-Typen, darunter drei neue (t4539, t6043, t11694) und ein Singleton (t779), die vier klonalen Komplexen zugeordnet wurden. Zwei tst- und drei luk-F/S-PV-tragende Stämme wurden identifiziert. Alle S. aureus Isolate waren mäßige Biofilmproduzenten mit verschiedenen Kombinationen der Biofilm- und icaR-Regulationsgene icaABCD. Der Nachweis genetisch unterschiedlicher S. aureus-Stämme und toxigener Stämme unterstreicht die Notwendigkeit einer verbesserten Überwachung resistenter und pathogener Stämme in Gesundheitseinrichtungen.
Schlüsselwörter
Biofilm, icaABCDR, Panton-Valentin-Leukozidin, HA-MRSA, Toxisches Schocksyndrom
Introduction
Burn and wound injuries are important causes of morbidity and death and are a common causes of hospitalization worldwide, necessitating both inpatient and outpatient care in many nations [1]. Bacterial infection accounts for significant morbidity and mortality in burn patients, and burn wound infection is the most common reason [2]. Burns and wounds provide a favorable site for bacterial multiplication and are more persistent, richer sources of infection than surgical wounds [3].
The bacteria that infect wounds and burn patients depend on multiple epidemiological factors. However, Staphylococcus aureus, a normal commensal in the nostrils and on skin, can be translocated to wounds and cause infectious processes [4]. Hence, S. aureus is one of the most frequently isolated pathogens in wounds and burns [5].
Healthcare-associated (HA)-MRSA is most frequently linked to a variety of infections in patients exposed to nosocomial settings. The emergence of community-associated (CA)-MRSA resulted in a significant shift in the epidemiology of MRSA isolates over the previous decades [4]. CA-MRSA can be distinguished by having distinct antimicrobial resistance patterns and molecular traits, although it is frequently identified by the lack of risk factors for HA-MRSA infections [6]. Generally, HA-MRSA typically harbours SCCmec I, II, and III, while CA-MRSA carries SCCmec IV or V [7]. Moreover, CA-MRSA isolates often carry the lukSF-PV genes that code for Panton-Valentine leukocidin (PVL), a cytolytic and toxic substance that has tropism for neutrophils [8].
The organism’s ability to spread rapidly through contact in the hospital ward poses a significant risk to managing burn and wound patients during hospital admission [9]. MRSA has been the center of concern due to its persistence and constant threat during the provision of healthcare services [10].
Moreover, healthcare workers (HCW) who frequently encounter sick individuals are at risk of contracting certain S. aureus strains from the patients and hospital environments. S. aureus is also often isolated from hospital curtains, surfaces, and equipment [11]. Hence, studying the nasal ecology of HCWs may provide greater insight into the potential transmission of pathogenic S. aureus strains in healthcare facilities. The treatment options for MRSA are becoming fewer by the day; this is associated with MRSA’s evolution into multi-drug resistance organisms, causing increased mortality around the globe [12]. Biofilm formation by MRSA worsens the situation by rendering it impenetrable, making the treatments more complex [13].
This study determined the frequency of S. aureus recovered from patients with burns and other wounds, as well as from healthcare workers at the Ahmadu Bello University Teaching Hospital, Zaria, Northern Nigeria. Moreover, the antimicrobial resistance (AMR) profile, biofilm formation capacity, biofilm and virulence genes, and lineages of the isolates were determined.
Methods
Study design and area
This cross-sectional study was conducted at Ahmadu Bello University Teaching Hospital Zaria, Kaduna State, on 206 burn- and wound patients (BWP) and 96 HCWs. The hospital is located in Shika, Zaria Local Government Area of Kaduna State, Nigeria. The 1000-bed capacity hospital serves as the main tertiary and reference hospital in the Northwest Geopolitical Zone. Approval (HREC number: ABUTHZ/HREC/W38/2020) was obtained from the Health Research Ethics Committee (HREC) of the Ahmadu Bello University Teaching Hospital, Zaria, before commencement of the study. All participants gave written informed consent before being recruited into this study.
Sample collection
The samples were collected aseptically. Swabs were taken from BWPs at sites with the highest deep-tissue exposure; the area was cleaned with sterile saline, after which the wound was swabbed. Moreover, a nasal swab was collected aseptically from HCWs (doctors, nurses, and hospital attendants) who were in contact with the BWPs. The samples were then transported to the Medical Microbiology Laboratory, ABUTH Zaria, for culture, bacterial isolation and identification.
Staphylococcus aureus identification
Each sample collected was cultured on Mannitol Salt agar (MSA) and then incubated for 24 hours at 37oC. The isolates were identified using the following conventional biochemical tests: gram staining, growth patterns on MSA (yellow colonies), hemolysis on blood agar, catalase test, rabbit plasma coagulase test (slide test), and DNAse test. At the same time, resistance against cefoxitin (30 µg) was considered a positive test for MRSA by subjecting each organism to a sensitivity test using the Kirby-Bauer method. The Clinical and Laboratory Standards Institute (CLSI) guideline was used to determine resistance [14]. S. aureus identification was performed using mass spectrometry Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) using Biotyper software (Bruker) and the standard extraction protocol recommended by the manufacturer, as previously described [15]. Briefly, from a pure culture grown for 24 hours at 37°C in Brain heart infusion (BHI) agar medium, a small portion of the bacterial colony was transferred to the 96-well metal plate and left to dry at room temperature. Thereafter, the wells were covered with 1 µL of an alpha-cyano-4 hydroxycinnamic acid matrix (HCCA; Bruker). For the calibration of the spectrometer, the protein profile of the E. coli strain DH5 peptide was used.
Extraction of bacterial DNA
For DNA extraction, the isolates were seeded on BHI agar and incubated for 24 h at 37°C. An isolated colony was suspended in 45 µL of sterile MiliQ water, then 5 µL of lysostaphin (1 mg/mL) (Sigma®) was added. The mixture was vortexed and incubated for 10 min at 37°C. Forty-five µL of sterile MiliQ water, 150 µL of Tris-HCl (0.1 M, pH 8) and 5 µL of proteinase K (2 mg/mL) (Sigma®) were added. This was vortexed and incubated for 10 min at 60°C. Finally, it was boiled for 5 min at 100°C and centrifuged at 12,000 rpm for 3 min. The DNA samples were stored at –20°C. The purity and concentration of the extracted DNA were measured using a NanoDrop UV-V is spectrophotometer (Thermo Fisher, USA).
Antibiotic susceptibility testing
Before the antibiotic susceptibility testing, the isolates were sub-cultured onto fresh nutrient agar slants incubated at 37°C for 24 hours. Suspensions were prepared from the sub-cultured isolates into clean, sterilized tubes using 0.5 McFarland’s standard. The isolates were then tested for their susceptibility to six antibiotics: penicillin G (10 IU), tetracycline (30 µg), erythromycin (15 µg), clindamycin (2 µg) doxycycline (30 µg), and levofloxacin (5 µg). The antibiotic discs (Oxoid, Thermo Fisher Scientific, Germany) were gently pressed to make sure they were in contact with the inoculated Mueller-Hinton agar surface, and the plates were incubated at 37°C for 24 hours. S. aureus ATCC 25923 was used as the control strain [16]. Zones of inhibition were measured to the nearest millimeter after incubation. The antibiotic breakpoints were determined using a chart adapted from the CLSI, 2022. Polymerase chain reaction (PCR) was conducted for the detection of the mecA gene from the Staphylococcus aureus isolates using the primers as presented in Table 1 [Tab. 1].
Table 1: Genes and primer sequences utilized in all PCRs 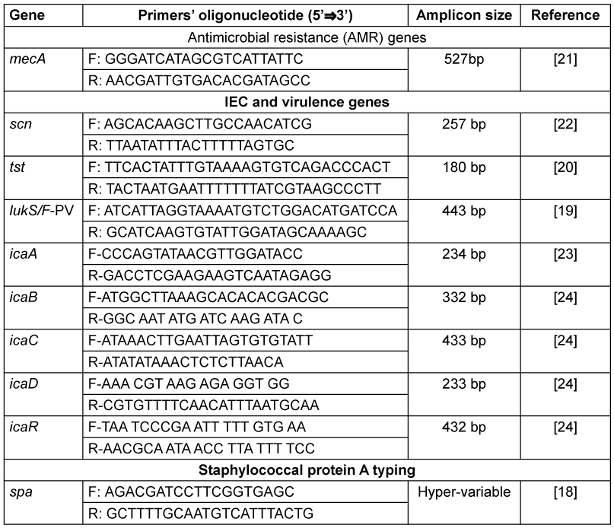
Phenotypic detection of biofilm formation
A tissue culture plate (TCP) was carried out using the method described by Ansari et al. [13]. Ten milliliters of tryptic soy broth (TSB) with 1% glucose in test tubes were inoculated with a loop-full of test organisms from overnight culture on nutrient agar. The test tube was incubated at 37°C for 24 hours, and a dilution of 1:100 with fresh medium was made. After gentle mixing, the 96-well flat- bottom TCPs were filled with 0.2 mL of the diluted cultures. Sterile broth was used to serve as the blank. The culture plates were incubated at 37°C for 24 hours. After incubation, the microtiter plates were gently tapped. The wells were washed with 0.2 mL of phosphate buffer saline at pH 7.2 four times to remove free-floating bacteria. In contrast, the biofilm that remained adhered to the wells’ walls and bottoms was fixed with 2% sodium acetate and stained with 0.1% crystal violet. Excess stain was washed with deionized water, and plates were thoroughly dried. An optical density (OD) of stained adherent biofilm was obtained with a microtiter plate reader at a wavelength of 570 nm. The experiment was performed in 3x3. The OD cut-off (ODc) was calculated, which is the average OD values of the non-inoculated medium (ODN) plus 3x standard deviation of the non-inoculated medium (3xSD). The degree of biofilm formation was determined using the following Equations:
Equation 1: OD<ODc: Non-biofilm producer.
Equation 2: OD≥ODc–OD<2xODc: Weak biofilm producer.
Equation 3: OD≥2xODc OD<4xODc: Moderate biofilm producer.
Equation 4: OD 4xODc: Strong biofilm producer.
Detection of biofilm-associated and regulatory genes
The isolates were investigated for the presence of genes associated with biofilm formation, namely: icaA, icaB, icaC, icaD, and icaR. Using the method described by Mottola et al [17], the amplification reactions contain a mixture of 12.5 µl of Supreme NZYTaq 2x Green Master Mix (NZYTech, Portugal), 2 µl of each primer (forward and reverse) (STAB VIDA Lda, Portugal) and 6.5µl of sterile water (water for molecular biology, NZYTech, Portugal). Two µl of the previously extracted DNA was added to the mixture, resulting in a total reaction volume of 25 µl. PCR amplification was conducted in a thermal cycler (MyCycler Thermal Cycler, Bio-Rad, Portugal).
Molecular typing (staphylococcal protein A)
All S. aureus strains were characterized by spa typing. After amplification of the hypervariable spa gene by PCR, Sanger sequencing of the amplicons was performed. For this, the variable fragment of the polymorphic region of the spa gene was amplified using forward and reverse primers (Table 1 [Tab. 1]) with the following cycling conditions: an initial denaturation at 95°C for 10 minutes followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 60°C for 60 seconds, extension at 72°C for 45 seconds, and final extension at 72°C for 10 minutes [18]. After PCR, the amplicons were visualized on agarose gel via electrophoresis and viewed on a gel dock. The PCR products were then sequenced, and the sequence obtained was analyzed with the Ridom® Staph-Type program (Ridom Gmbh; https://www.spaserver.ridom.de), which automatically assigns the spa type according to the repetitions detected. The clonal complex of the isolates was assigned according to the spa type, when possible.
Detection of S. aureus toxigenic genes
The S. aureus isolates were tested for the presence of lukS-PV-lukF-PV under the following cycling conditions: initial denaturation at 94°C for 5 minutes followed by 30 cycles of denaturation at 94°C for 30 seconds annealing at 55°C for 30 seconds, extension at 72°C for 1 minute and final extension at 72ºC for 10 minutes [19]. Moreover, all the strains were tested for the presence of the tst gene with a cycling condition of initial denaturation at 94°C for 5 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72ºC for 1 minute and final extension at 72°C for 10 minutes [20].
Detection of staphylococcal complement inhibitor (scn) gene
The MRSA and MSSA isolates were tested for the presence of the staphylococcal complement inhibitor (scn) gene with the following cycling conditions: initial denaturation at 95°C for 3 minutes, a series of denaturation at 94°C for 30 seconds, annealing at 53°C for 30 seconds for 30 cycles followed by extension at 72ºC for 2 minutes and final extension at 72ºC for 6 min.
Results
Demographic characteristics of the study participants
Of the three hundred participants, 206 (68.7%) were BWPs and 94 (31.3%) were HCWs. Males comprised 147 (49.0%) and females 153 (51.0%) of the study population. The median age of the study participants was 30.5 (IQR: 22–40) years. According to age categorization, the age group of 21–30 years was most highly represented with 90 (30.3%) participants, and the age group of 1–10 years was the least among the patients (4.7%). On the other hand, the study participants who were HCWs comprised 12 (4.0%) doctors, 52 (17.3%) nurses and 30 (10%) health assistants (Table 2 [Tab. 2]).
Table 2: Socio-demographic characteristics of the participants 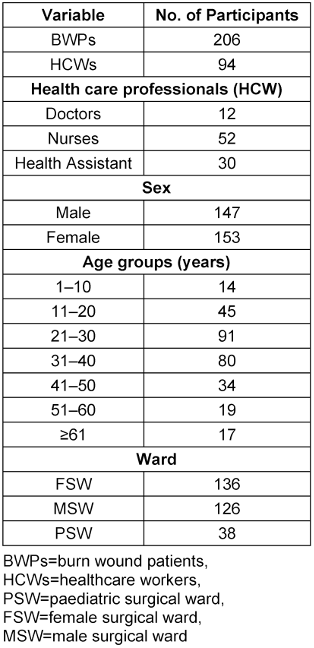
Prevalence of S. aureus among study participants
A total of 68 (23%) samples were culture-positive on the Mannitol Salt Agar medium, and one isolate per sample was selected. After identification, only nine (4.4%) and five (5.3%) isolates from BWPs and HCWs consisted of S. aureus, respectively (Figure 1 [Fig. 1]). Eight (8) were S. scuiri, while S waneri, S eqourum and S. heamolyticus were found in one sample each. The mecA gene was detected in 7 (50%) (Table 3 [Tab. 3]). Of this, MRSA strains were detected in four BWPS (1.9%) and three HCWs (3.2%).
Figure 1: Frequency of S. aureus and MRSA carriage or infection among the study participants 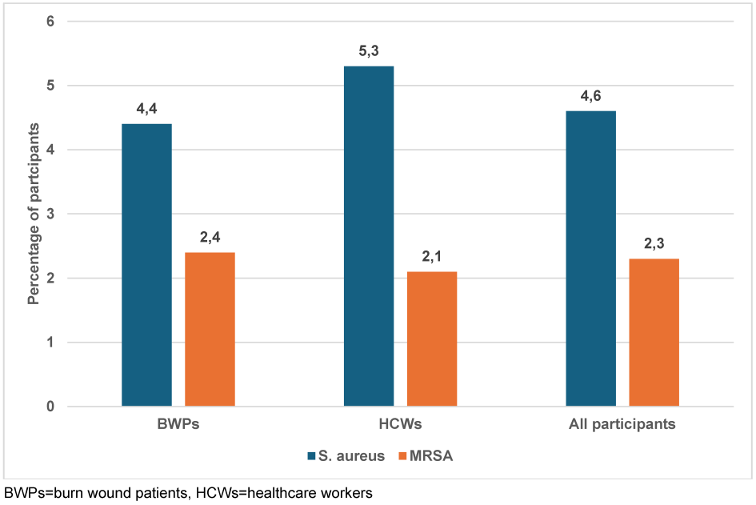
Table 3: Molecular typing and antimicrobial resistance profile of the 14 S. aureus isolates from the study participants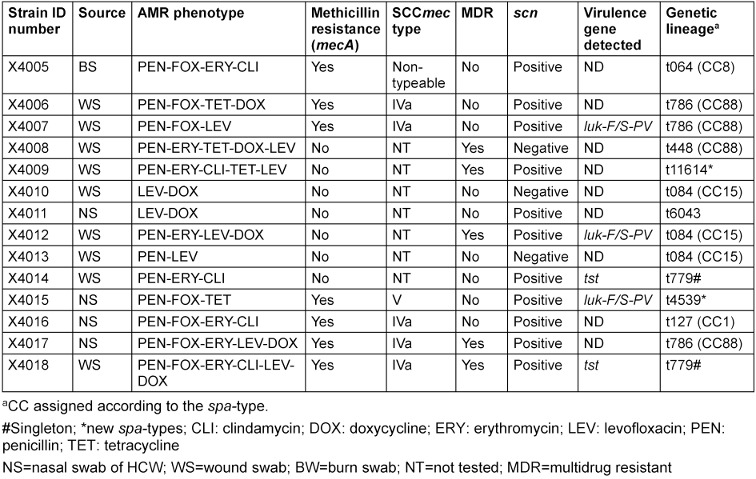
Antimicrobial resistance and virulence profile of the S. aureus strains
Seven S. aureus isolates (50%) were mecA-positive, two from BWPs and 5 from HCWs (associated with SCCmec types IVa and V), while 35.7% presented a multidrug resistance (MDR) phenotype. The following AMR phenotypes were obtained among S. aureus recovered from both HCWs and BWPs: penicillin (100%), levofloxacin (64.3%), doxycycline (50%), tetracycline (28.6%), erythromycin-clindamycin-constitutive (35.7%) and erythromycin (21.4%) (Table 3 [Tab. 3]).
The S. aureus isolates belonged to 11 different spa types, including three new (t4539, t6043 and t11694) and one singleton (t779), and were assigned to four clonal complexes (CC1, CC8, CC15 and CC88). Of these, CC88 (three MRSA and one MSSA) was the predominant complex (28.6%). The S. aureus lineages t064 and t127 came exclusively from HCWs, while t779 and t4539 were found in BWPs. Two tst-carrying strains (MRSA-t779 and MSSA-t779) and three luk-F/S-PV carrying strains (one MRSA-CC88-t786, MRSA-CC15-t084, MSSA-t4539) were identified. Some of the isolates (21.4%) were negative for the scn gene (human adaptation marker). All the S. aureus isolates were moderate biofilm producers with diverse combinations of the icaABCD biofilm and icaR regulatory genes (Table 4 [Tab. 4]).
Table 4: Molecular characteristics and virulence genes of the 14 S. aureus strains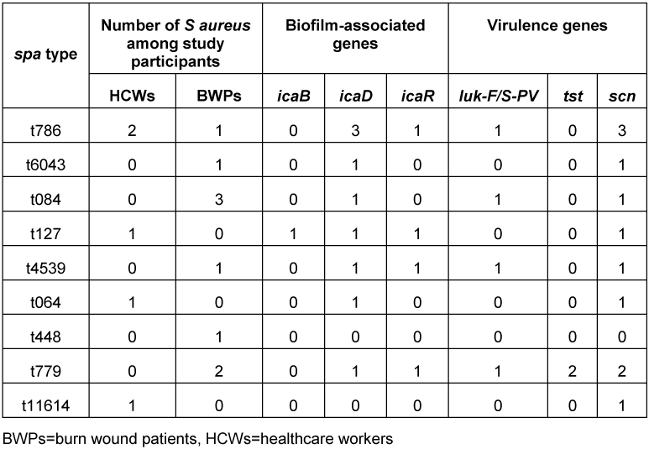
Discussion
Methicillin-resistant S. aureus has been the center of concern due to its persistence and and constant threat during the provision of healthcare services [10]. Biofilm formation by MRSA worsens the situation by rendering it impenetrable, making treatment more complex [13]. Burn injuries are a significant health concern, particularly in resource-limited settings. Studying the characteristics and experiences of both patients and healthcare workers can contribute to better burn-care practices and interventions [21].
The overall prevalence of S. aureus was 4.6%, while that among HCWs and BWPs was 5.3% and 4.3% respectively, which is consistent with the finding of Gajdács et al. [22]. The lower sensitivity of the phenotypic method for identification of S. aureus could be due to the similarity of bacterial cultural features of S. aureus and other non-aureus staphylococci, such as S. haemolyticus and S. xylosus [23]. Moreover, the agglutination method for the detection of coagulase enzyme could produce a false positive reaction [24], [25].
The overall prevalence of MRSA after mecA detection among recovered S. aureus was high (50%). This was lower than that obtained by Angel et al. [26] in Abuja Nigeria, who reported 93.8% mecA-positive isolate from patients, and higher than that obtained by Joshua et al. [27], who reported 15% in Zaria. Specifically, the low prevalence of S. aureus and MRSA identified from the BWPs in this study suggests that bacterial species other than S. aureus could have predominated in wound/burn infections [28]. In contrast, in the HCWs, the low frequencies of S. aureus/MRSA could be an indication of good infection control practices in the hospital.
All MRSA isolates found in this study were phenotypically moderate biofilm producers, which was corroborated by genotypic detection of either of the biofilm-associated genes (ica) in all MRSA isolates. These findings are similar to the findings of Leshem et al. [29], who compared the ability to detect biofilm production of MSSA and MRSA by TCP and Congo Red methods. In addition, Silva et al. [30] reported agreement between biofilm detection through the TCP method and the detection of biofilm-associated genes. Similarly, Oche et al. [31] in Kano Nigeria reported a 94% detection rate of biofilm-producing S. aureus with a 100% biofilm-production rate of MRSA among orthopedic patients. The high rate of biofilm formation by MRSA could be linked to impaired wound healing due to the association of biofilm production and virulence of MRSA [32].
The tst gene that encodes toxic shock syndrome was detected in two isolates obtained from BWPs (one MRSA-t779 and one MSSA-t779). This is similar to finding of a study that reported the predominance of tst-producing S. aureus-spa type t779 lineage [33]. Contrary to our findings, Soltani et al. [34] reported that up to 18% of MRSA isolated from hospital settings carries the tst gene. However, luk-F/S-PV-carrying S. aureus strains were detected from both HCWs and BWPs. In this regard, the luk-F/S-PV-carrying MSSA strain was from only a BWP, while luk-F/S-PV-carrying MRSA strains were detected from a BWP and an HCW. These findings are similar to those previously reported by Joshua et al. [27] in Zaria and [35] in Abuja, in which they reported that about 10% of the S. aureus isolates carried the luk-F/S-PV gene. It is important to remark that all the participants who harbored the tst and luk-F/S-PV genes were BWPs. This could be because both virulence genes are predominantly associated with CA-MRSA, which suggests that patients might have contacted the organism even before admission [36]. All the toxigenic MRSA strains had SCCmec types (IVc and V), which denoted community acquisition.
Unlike tst and luk-F/S-PV, the scn gene (a host adaptation marker) was reported from both MRSA and MSSA. The presence of scn in most of the isolates suggests a potentially human origin [37]. The strains that were scn-negative could be due to loss of the sa3 prophage [38], which suggests animal origin and highlights potential zoonotic infection [39].
Diverse S. aureus lineages were identified, but the lineages CC15 and CC88 predominated in all participants. The MRSA-CC88 is termed the African Clone [40] and appears circulate widely in Nigeria [41], [42], [43]. It has been shown that the CC88 lineage is predominantly PVL-positive and spreads globally in hospital facilities [44], [45], [46], [47]. In this regard, the only MSSA-CC88 strain from the present study is scn-negative, which could support the previous findings.
The lineage CC15 is ubiquitous and widely described in the literature, but these isolates are mostly MSSA and are often nasal colonizers [48]. Hence, the presence of this lineage in the BWPs indicates the translocation from the nose to the wound surfaces.
Concerning the AMR profiles, relatively high resistance to fluoroquinolones, macrolide-lincosamide-streptogramins-B, and tetracycline were found. The macrolide and levofloxacin resistance was not surprising, as they among the top classes of antibiotics frequently prescribed in Nigeria [49]. However, tetracycline is a major antimicrobial agent of choice against Staphylococcus in both human and veterinary medicine [50]. Thus, resistance to these categories of antibiotics might be associated with high selective pressure due to their frequent use.
Limitation
Swabbing is not the gold standard for sampling from wounds; for instance, tissue biopsy or the Levine technique are more effective. Because of this, not all S. aureus could be recovered by swabbing. Other wound-associated bacterial pathogens could also be found in wound/burn patients. However, the focus of the present study was to investigate the potential contamination (nosocomial transmission) in either direction of wound patients or healthcare workers by the major nasal colonizer S. aureus. Thus, other bacteria were excluded.
Conclusions
A low frequency of S. aureus with less biofilm-producing ability was obtained. However, most of the isolates presented an MDR phenotype; a significant number of them were toxigenic. The results do not indicate a nosocomial event. However, the detection of diverse S. aureus lineages, resistance to first-line clinical antibiotics, and the ability of the virulent MRSA isolates highlight the need for improved surveillance of resistant and pathogenic strains in healthcare facilities.
Notes
Competing interests
The authors declare that they have no competing interests.
Data Availability Statement
All the data derived from this study have been presented in this article. However, additional information may be requested from the corresponding author (Dr. Idris Nasir Abdullahi).
Authors’ ORCID
- Umar K: 0000-0003-2500-2822
- Abdullahi IN: 0000-0002-5511-1272
- Usman Y: 0000-0003-3972-5351
- El-fulaty Ahmad A: 0000-0003-1941-8346
- Torres C: 0000-0003-3709-1690
Funding
This study was funded by TetFund Nigeria through an institutional-based research grant (grant number: TETF/DR&D/UNI/ZARIA/IBR/2020/VOL.1/4).
References
[1] Zhou S, Xiao S, Wang X, Wang X, Han L. Risk Factors and Pathogens of Wound Infection in Burn Inpatients from East China. Antibiotics (Basel). 2023 Sep;12(9:1432. DOI: 10.3390/antibiotics12091432[2] El Hamzaoui N, Barguigua A, Larouz S, Maouloua M. Epidemiology of burn wound bacterial infections at a Meknes hospital, Morocco. New Microbes New Infect. 2020 Nov;38:100764. DOI: 10.1016/j.nmni.2020.100764
[3] Alebachew T, Yismaw G, Derabe A, Sisay Z. Staphylococcus aureus burn wound infection among patients attending yekatit 12 hospital burn unit, addis ababa, ethiopia. Ethiop J Health Sci. 2012 Nov;22(3):209-13.
[4] Abdullahi IN, Lozano C, Ruiz-Ripa L, Fernández-Fernández R, Zarazaga M, Torres C. Ecology and Genetic Lineages of Nasal and MRSA Carriage in Healthy Persons with or without Animal-Related Occupational Risks of Colonization: A Review of Global Reports. Pathogens. 2021 Aug 8;10(8):1000. DOI: 10.3390/pathogens10081000
[5] Hong X, Zhou S, Dai X, Xie D, Cai Y, Zhao G, Li B. Molecular typing and characterization of isolates from burn wound infections in Fujian, China. Front Microbiol. 2023;14:1236497. DOI: 10.3389/fmicb.2023.1236497
[6] Siddiqui AH, Koirala J. Methicillin-resistant Staphylococcus aureus, 2023. In: StatPearls. Treasure Island (FL): StatPearls Publishing; 2024 Jan [last update: 2023 Apr 2].
[7] Moosavian M, Shahin M, Navidifar T, Torabipour M. Typing of staphylococcal cassette chromosome encoding methicillin resistance in isolates in Ahvaz, Iran. New Microbes New Infect. 2018 Jan;21:90-4. DOI: 10.1016/j.nmni.2017.11.006
[8] Otter JA, Kearns AM, French GL, Ellington MJ. Panton-Valentine leukocidin-encoding bacteriophage and gene sequence variation in community-associated methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2010 Jan;16(1):68-73. DOI: 10.1111/j.1469-0691.2009.02925.x
[9] Emaneini M, Beigverdi R, van Leeuwen WB, Rahdar H, Karami-Zarandi M, Hosseinkhani F, Jabalameli F. Prevalence of methicillin-resistant Staphylococcus aureus isolated from burn patients in Iran: A systematic review and meta-analysis. J Glob Antimicrob Resist. 2018 Mar;12:202-6. DOI: 10.1016/j.jgar.2017.10.015
[10] Eftekhar R, Rezaee R, Azad M, Azimi H, Goudarzi H, Goudarzi M. Distribution of Adhesion and Toxin Genes in Staphylococcus aureus Strains Recovered From Hospitalized Patients Admitted to the ICU. Arch Pediatr Infect Dis. 2017;5(1):e39349. DOI: 10.5812/pedinfect.39349
[11] Shek K, Patidar R, Kohja Z, Liu S, Gawaziuk JP, Gawthrop M, Kumar A, Logsetty S. Rate of contamination of hospital privacy curtains in a burns/plastic ward: A longitudinal study. Am J Infect Control. 2018 Sep;46(9):1019-21. DOI: 10.1016/j.ajic.2018.03.004
[12] Fasihi Y, Kiaei S, Kalantar-Neyestanaki D. Characterization of SCCmec and spa types of methicillin-resistant Staphylococcus aureus isolates from health-care and community-acquired infections in Kerman, Iran. J Epidemiol Glob Health. 2017 Dec;7(4):263-7. DOI: 10.1016/j.jegh.2017.08.004
[13] Ansari MA, Khan HM, Khan AA, Cameotra SS, Alzohairy MA. Anti-biofilm efficacy of silver nanoparticles against MRSA and MRSE isolated from wounds in a tertiary care hospital. Indian J Med Microbiol. 2015;33(1):101-9. DOI: 10.4103/0255-0857.148402
[14] Goudarzi M, Kobayashi N, Hashemi A, Fazeli M, Navidinia M. Genetic Variability of Methicillin Resistant Strains Isolated from Burns Patients. Osong Public Health Res Perspect. 2019 Jun;10(3):170-6. DOI: 10.24171/j.phrp.2019.10.3.08
[15] Torres-Sangiao E, Leal Rodriguez C, García-Riestra C. Application and Perspectives of MALDI-TOF Mass Spectrometry in Clinical Microbiology Laboratories. Microorganisms. 2021 Jul;9(7):. DOI: 10.3390/microorganisms9071539
[16] Kouadri F. In vitro antibacterial and antifungal activities of the Saudi Lawsonia inermis extracts against some nosocomial infection pathogens. J Pure Appl Microbiol. 2018;12(1):281-6.
[17] Mottola C, Matias CS, Mendes JJ, Melo-Cristino J, Tavares L, Cavaco-Silva P, Oliveira M. Susceptibility patterns of Staphylococcus aureus biofilms in diabetic foot infections. BMC Microbiol. 2016 Jun;16(1):119. DOI: 10.1186/s12866-016-0737-0
[18] Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003 Dec;41(12):5442-8. DOI: 10.1128/JCM.41.12.5442-5448.2003
[19] Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999 Nov;29(5):1128-32. DOI: 10.1086/313461
[20] Yamaguchi T, Nishifuji K, Sasaki M, Fudaba Y, Aepfelbacher M, Takata T, Ohara M, Komatsuzawa H, Amagai M, Sugai M. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect Immun. 2002 Oct;70(10):5835-45. DOI: 10.1128/IAI.70.10.5835-5845.2002
[21] Fadeyibi IO, Raji MA, Ibrahim NA, Ugburo AO, Ademiluyi S. Bacteriology of infected burn wounds in the burn wards of a teaching hospital in Southwest Nigeria. Burns. 2013 Feb;39(1):168-73. DOI: 10.1016/j.burns.2012.02.005
[22] Gajdács M, Ábrók M, Lázár A, Burián K. Comparative Epidemiology and Resistance Trends of Common Urinary Pathogens in a Tertiary-Care Hospital: A 10-Year Surveillance Study. Medicina (Kaunas). 2019 Jul;55(7):356. DOI: 10.3390/medicina55070356
[23] Thakur P, Nayyar C, Tak V, Saigal K. Mannitol-fermenting and Tube Coagulase-negative Staphylococcal Isolates: Unraveling the Diagnostic Dilemma. J Lab Physicians. 2017;9(1):65-6. DOI: 10.4103/0974-2727.187926
[24] Kumurya A. Malfunction of agglutination test to identify methicillin-resistant Staphylococcus aureus strains (MRSA). Int J Bioinf Biomedical Engin. 2015;1:1-6.
[25] Abalkhail A, Elbehiry A. Methicillin-resistant Staphylococcus aureus in diabetic foot infections: protein profiling, virulence determinants, and antimicrobial resistance. Appl Sci. 2022;12(21):10803. DOI: 10.3390/app122110803.
[26] Angel OD, Kadarko PS, Muazu JS, Bassey BE, Helma AR, Haruna NI, Boyi NY. Antimicrobial resistance profile and molecular detection of mecA gene in methicillin resistant Staphylococcus aureus from patients in selected general hospitals in Abuja municipal, Nigeria. GSC Biol Pharmac Sci. 2019 Jun 30;7(3):93-106. DOI: 10.30574/gscbps.2019.7.3.0090
[27] Joshua IA, Giwa FJ, Kwaga JKP, Kabir J, Owolodun OA, Umaru GA, Habib AG. Phenotypic Characterisation of Staphylococcus aureus Isolated from Patients in Healthcare Institutions in Zaria Metropolis, Kaduna State, Nigeria. West Afr J Med. 2022 Nov 30;39(11):1148-55.
[28] Li Z, Xie J, Yang J, Liu S, Ding Z, Hao J, Ding Y, Zeng Z, Liu J. Pathogenic Characteristics and Risk Factors for ESKAPE Pathogens Infection in Burn Patients. Infect Drug Resist. 2021;14:4727-38. DOI: 10.2147/IDR.S338627
[29] Leshem T, Schnall BS, Azrad M, Baum M, Rokney A, Peretz A. Incidence of biofilm formation among MRSA and MSSA clinical isolates from hospitalized patients in Israel. J Appl Microbiol. 2022 Aug;133(2):922-9. DOI: 10.1111/jam.15612
[30] Silva V, Almeida L, Gaio V, Cerca N, Manageiro V, Caniça M, Capelo JL, Igrejas G, Poeta P. Biofilm Formation of Multidrug-Resistant MRSA Strains Isolated from Different Types of Human Infections. Pathogens. 2021 Jul 30;10(8):970. DOI: 10.3390/pathogens10080970
[31] Oche DA, Abdulrahim U, Oheagbulem AS, Olayinka BO. Isolation of biofilm producing methicillin-resistant Staphylococcus aureus from hospitalized orthopaedic patients in Kano State, Nigeria. Niger J Basic Appl Sci. 2020; 28(1):66-74. DOI: 10.4314/njbas.v28i1.9.
[32] Serray B, Oufrid S, Hannaoui I, Bourjilate F, Soraa N, Mliji M, Sobh M, Hammoumi A, Timinouni M, El Azhari M. Genes encoding adhesion factors and biofilm formation in methicillin-resistant Staphylococcus aureus in Morocco. J Infect Dev Ctries. 2016 Aug;10(8):863-9. DOI: 10.3855/jidc.8361
[33] Mama OM, Morales L, Ruiz-Ripa L, Zarazaga M, Torres C. High prevalence of multidrug resistant S. aureus-CC398 and frequent detection of enterotoxin genes among non-CC398 S. aureus from pig-derived food in Spain. Int J Food Microbiol. 2020 May;320:108510. DOI: 10.1016/j.ijfoodmicro.2020.108510
[34] Soltani M, Hajikhani B, Zamani S, Haghighi M, Hashemi A, Nasiri MJ, Nasiri M, Dadashi M, Pourhossein B, Goudarzi M. Molecular characterization of Staphylococcus aureus strains isolated from hospitalized patients based on coagulase gene polymorphism analysis: High frequency of vancomycin-intermediate S. aureus and the emergence of coagulase type II in Iran. Gene Rep. 2021 Jun 1;(23):1-7. DOI: 10.1016/J.GENREP.2021.101078.
[35] Orji OL, Olayinka BO, Afolabi B, Ejikeugwu CP. Molecular detection of panton-valentine leukocidin (PVL) toxins in clinical isolates of Staphylococcus aureus from Maitama district hospital, Abuja, Nigeria. J Med Microbiol Diagn. 2016;5:240. DOI: 10.4172/2161-0703.1000240.
[36] Dad V, Ahmadrajabi R, Esfahani S, Saffari F. Vergleichende Studie zu Staphylococcus aureus bei Brandverletzten und medizinischem Personal in einem Verbrennungszentrum, Yazd, Iran Comparative study of Staphylococcus aureus from burn patients and healthcare workers in a burn center, Yazd, Iran. Wien Med Wochenschr. 2022 Sep;172(11-12):256-60. DOI: 10.1007/s10354-021-00863-5
[37] Chenouf NS, Mama OM, Messaï CR, Ruiz-Ripa L, Fernández-Fernández R, Carvalho I, Torres C. Detection of methicillin-resistant coagulase-negative staphylococci and PVL/mecA genes in cefoxitin-susceptible Staphylococcus aureus (t044/ST80) from unpasteurized milk sold in stores in Djelfa, Algeria. J Dairy Sci. 2021; 104(3):2684-92. DOI: 10.3168/jds.2020-19270.
[38] Abdullahi IN, Lozano C, Saidenberg ABS, Latorre-Fernández J, Zarazaga M, Torres C. Comparative review of the nasal carriage and genetic characteristics of Staphylococcus aureus in healthy livestock: Insight into zoonotic and anthroponotic clones. Infect Genet Evol. 2023 Apr;109:105408. DOI: 10.1016/j.meegid.2023.105408
[39] Lozano C, Fernández-Fernández R, Ruiz-Ripa L, Gómez P, Zarazaga M, Torres C. Human mecC-Carrying MRSA: Clinical Implications and Risk Factors. Microorganisms. 2020 Oct 20;8(10):1615. DOI: 10.3390/microorganisms8101615
[40] Shittu AO, Adesoji T, Udo EE. DNA microarray analysis of Staphylococcus aureus from Nigeria and South Africa. PLoS One. 2021;16(7):e0237124. DOI: 10.1371/journal.pone.0237124
[41] Okorie-Kanu OJ, Anyanwu MU, Ezenduka EV, Mgbeahuruike AC, Thapaliya D, Gerbig G, Ugwuijem EE, Okorie-Kanu CO, Agbowo P, Olorunleke S, Nwanta JA, Chah KF, Smith TC. Molecular epidemiology, genetic diversity and antimicrobial resistance of Staphylococcus aureus isolated from chicken and pig carcasses, and carcass handlers. PLoS One. 2020;15(5):e0232913. DOI: 10.1371/journal.pone.0232913
[42] Odetokun IA, Adetona MA, Ade-Yusuf RO, Adewoye AO, Ahmed AN, Ghali-Mohammed I, Al Mustapha AI, Fetsch A. Staphylococcus aureus contamination of animal-derived foods in Nigeria: a systematic review, 2002—2022. Food Safety Risk. 2023;10(6):. DOI: 10.1186/s40550-023-00106-y
[43] Mrochen DM, Grumann D, Schulz D, Gumz J, Trübe P, Pritchett-Corning K, Johnson S, Nicklas W, Kirsch P, Martelet K, Brandt JVD, Berg S, Bröker BM, Wiles S, Holtfreter S. Global spread of mouse-adapted Staphylococcus aureus lineages CC1, CC15, and CC88 among mouse breeding facilities. Int J Med Microbiol. 2018 Aug;308(6):598-606. DOI: 10.1016/j.ijmm.2017.11.006
[44] Senok A, Somily AM, Nassar R, Garaween G, Kim Sing G, Müller E, Reissig A, Gawlik D, Ehricht R, Monecke S. Emergence of novel methicillin-resistant strains in a tertiary care facility in Riyadh, Saudi Arabia. Infect Drug Resist. 2019;12:2739-46. DOI: 10.2147/IDR.S218870
[45] Alfouzan WA, Boswihi SS, Udo EE. Methicillin-Resistant (MRSA) in a Tertiary Care Hospital in Kuwait: A Molecular and Genetic Analysis. Microorganisms. 2023 Dec;12(1):. DOI: 10.3390/microorganisms12010017
[46] Tkadlec J, Le AV, Brajerova M, Soltesova A, Marcisin J, Drevinek P, Krutova M. Epidemiology of Methicillin-Resistant Staphylococcus aureus in Slovakia, 2020 - Emergence of an Epidemic USA300 Clone in Community and Hospitals. Microbiol Spectr. 2023 Aug;11(4):e0126423. DOI: 10.1128/spectrum.01264-23
[47] Gordon NC, Pichon B, Golubchik T, Wilson DJ, Paul J, Blanc DS, Cole K, Collins J, Cortes N, Cubbon M, Gould FK, Jenks PJ, Llewelyn M, Nash JQ, Orendi JM, Paranthaman K, Price JR, Senn L, Thomas HL, Wyllie S, Crook DW, Peto TEA, Walker AS, Kearns AM. Whole-Genome Sequencing Reveals the Contribution of Long-Term Carriers in Staphylococcus aureus Outbreak Investigation. J Clin Microbiol. 2017 Jul;55(7):2188-97. DOI: 10.1128/JCM.00363-17
[48] Abdullahi IN, Fernández-Fernández R, Juárez-Fernández G, Martínez-Álvarez S, Eguizábal P, Zarazaga M, Lozano C, Torres C. Wild Animals Are Reservoirs and Sentinels of and MRSA Clones: A Problem with "One Health" Concern. Antibiotics (Basel). 2021 Dec;10(12):. DOI: 10.3390/antibiotics10121556
[49] Abubakar U. Antibiotic use among hospitalized patients in northern Nigeria: a multicenter point-prevalence survey. BMC Infect Dis. 2020 Jan;20(1):86. DOI: 10.1186/s12879-020-4815-4
[50] Abdullahi IN, Lozano C, Zarazaga M, Simón C, Höfle U, Sieber RN, Latorre-Fernández J, Stegger M, Torres C. Comparative genomics of Staphylococcus aureus strains from wild birds and pig farms elucidates levels of mobilomes, antibiotic pressure and host adaptation. J Glob Antimicrob Resist. 2024 Mar;36:142-50. DOI: 10.1016/j.jgar.2023.12.003
[51] Poulsen AB, Skov R, Pallesen LV. Detection of methicillin resistance in coagulase-negative staphylococci and in staphylococci directly from simulated blood cultures using the EVIGENE MRSA Detection Kit. J Antimicrob Chemother. 2003 Feb;51(2):419-21. DOI: 10.1093/jac/dkg084
[52] van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J Bacteriol. 2006 Feb;188(4):1310-5. DOI: 10.1128/JB.188.4.1310-1315.2006
[53] Boopathy R. Presence of Methicillin Resistant Staphylococcus aureus (MRSA) in sewage treatment plant. Bioresour Technol. 2017 Sep;240:144-8. DOI: 10.1016/j.biortech.2017.02.093
[54] Ziebuhr W, Krimmer V, Rachid S, Lössner I, Götz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999 Apr;32(2):345-56. DOI: 10.1046/j.1365-2958.1999.01353.x




