Automated metadata transformation in a medical data integration center: Implementation of an algorithm and standardized quality analysis
Caroline Bönisch 11 Department of Medical Informatics, Medical Data Integration Center, University Medical Center Göttingen, Germany
Abstract
This study presents a novel approach to metadata management, while focusing on the development of an automated, microservice-based infrastructure at the University Medical Center Göttingen’s Medical Data Integration Center (UMG-MeDIC). Given the critical role of high-quality metadata in supporting reliable clinical research and data interoperability, this research addresses the challenges of metadata extraction, storage, and quality assurance across multiple data formats used in healthcare. Through a mixed-methods, single-case design, the metadata framework was developed, adopting a convergence format that integrates metadata from standards such as CDISC, OMOP, openEHR, and FHIR. This format enables consistent metadata representation and accommodates missing values, thus preserving data integrity and supporting FAIR principles. Key quality metrics, including completeness, consistency, and relevance, were defined and operationalized to assess metadata reliability systematically. The microservice architecture used enhances scalability and adaptability, demonstrating a replicable model for other data integration centers (DICs). This work contributes a scalable framework for metadata management, with potential for further application.
Keywords
metadata, quality, interoperability, reliability
Introduction
Within the constantly changing healthcare environment, the incorporation and administration of clinical data are pivotal for delivering high-quality patient care, streamlining operational procedures, and facilitating clinical research [1]. In order to achieve high-quality healthcare, the usefulness of clinical data collected depends largely on its quality [2], [3].
Evidence-based medicine, enhancing the quality of diagnosis and treatment, is dependent on the clinicians’ practical experience and robust medical knowledge derived from collaborative research across multiple institutions [4].
In addition, the Food and Drug Administration (FDA) has designated real-world data (RWD) as an additional pillar of complementary information to the randomized clinical trial (RCT) in evidence-based medicine [5]. RWD encompasses all information that is gathered from routine clinical practices provided within a healthcare service context to the patient. These data are then repurposed beyond their original intent of collection.
Subsequently, with the introduction of the FAIR Guiding Principles by Wilkinson et al. [6], data proprietors received directives aimed at ensuring that the collected data is both discoverable and accessible, exhibits interoperability, and is amenable to reuse. The importance of metadata, as an integral part of data, is highlighted across all FAIR principles. The significance of metadata, as primary cue and first information access to datasets, is omnipresent throughout every stage of the data processing of medical data [6].
Metadata are essentially data that provide information about other data. They describe information objects in terms of their source, creation type, structure, status, level, and meaning. An information object may be a single data point, such as a coded value or an instance identifier, a series of dates, or even an entire database with multiple interdependencies [7]. This research follows the definition of metadata by Canham and Ohmann with the proposed differentiation between intrinsic and provenance metadata, which complements the FAIR principles [8].
An additional unique aspect of the FAIR principles includes that data must be centralized in order to provide the basis of the FAIRification process. For this purpose, it is necessary to maintain a centralized data collection point within the clinical area [9].
The responsibility for this task within the University Medical Center Göttingen lies with the Medical Data Integration Center (UMG-MeDIC). The UMG-MeDIC consolidates data from a hospital of maximum care and operates on data originating from controlled studies and clinical care data, commonly referred to as RWD. The UMG-MeDIC is moreover the first point of contact to give researchers access to the merged data pool of clinical data.
The problem that arises in the provision of metadata is the qualitative heterogeneity of the underlying clinical data. The objectives of this work therefore include investigating and assessing the data quality of clinical data, as well as providing the necessary information on quality for researchers within metadata.
Methods
Building on the objective of this study, meta-information on different data structures has to be made available to researchers in order to release the information independently of data format restrictions.
The given project is structured as topdown research, adopting an inductive approach. Since the work heavily depends on specific data and metadata, drawing conclusions based on the data’s structure and the configuration of the respective metadata, the research can be characterized as confirmatory.
In line with this inductive approach, the research employs a mixed methodology, integrating both quantitative and qualitative research. The UMG-MeDIC’s data and metadata are examined in terms of their compatibility with specified data formats used in healthcare, as well as the correlation between metadata and data quality in clinical care data. The open-ended research questions outlined in Table 1 [Tab. 1] enhance the qualitative aspect of this research, while the numerical assignment and assessment of metadata align with a quantitative approach.

In accordance with characteristics proposed by Benbasat et al. [10], the envisioned strategy is characterized as a case research. The research adopts a single-case research design, given that the described situation was previously inaccessible of scientific investigations and only became possible with the establishment of a MeDIC at the University Medical Center Göttingen.
In the case of research studies, data analysis relies on diverse methods of data collection that involve combining one or more sources to generate generalizable findings. According to Yin [11], documentation, archival records, interviews, direct observation, and physical artifacts constitute key data collection methods in case research. This work primarily focuses on documentation, direct observation, and physical artifacts to gather a comprehensive dataset related to the specific research issue
The cumulative elements in form of peer-reviewed publications, as part of the thesis, refer to the methodologies described above. The division into separate parts allowed for the differentiated use of various research methods (mixed method approach) within the overarching case research. Figure 1 [Fig. 1] shows an overview of the methodologies used within the respective publications.
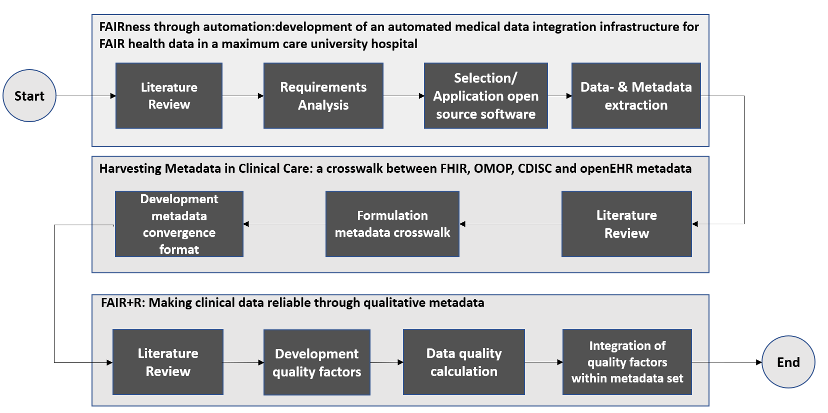
Within the first part, setting up the MeDIC at the UMG, requirements for the implementation of the UMG-MeDIC were identified on the basis of a literature review to derive various approaches for the development of a FAIRified data infrastructure. The tools and approaches used to set up the UMG-MeDIC for data processing were then defined. Finally, the defined objectives were integrated into the UMG-MeDIC framework as stated by Parciak et al. [12].
By defining and setting up the UMG-MeDIC framework, the preliminary work on data and metadata extraction was made. Therefore, metadata harvesting, a process of gathering metadata from various data storages, archives or repositories, with the intention of consolidating the information into a generic database schema, was conducted. Metadata that were harvested and analyzed in the process included (among others) metadata about the data lifecycle state (e.g. creation, processing, analysis, preservation, access, reuse), metadata about the data usage license and metadata about the source system, where the data was originally created. In order to be able to design a corresponding generic database schema for storing the metadata, various data formats for storing medical data were examined [13].
While focusing on the examination of the data format specifications of Clinical Data Interchange Standards Consortium (CDISC), Observational Medical Outcomes Partnership (OMOP), open Electronic Health Record (openEHR), and Fast Healthcare Interoperability Ressources (FHIR), corresponding metadata items for each data format were extracted and compared from the documentation of each data format. Based on the identified metadata a priorization was conducted, with regards to the requirements inheritant of the UMG-MeDIC. After the priorization, it became apparent that not one single, but a specifically developed convergence format fulfilled the requirements on metadata within the UMG-MeDIC as shown in Bönisch et al. [13]. The convergence format demonstrates how metadata items from different formats can be incorporated, preventing the loss of information by providing metadata items in the target format, even if they are not present in the source data format. The convergence format offers the best solution for maintaining the structure of the format by generating the required items during the transformation process and populating them with NULL values if the source format does not provide any input values [13].
Furthermore the research aimed to determine and provide information on the quality of clinical (meta)data, based on the proposed quality factors: completeness, consistency, correctness, correspondence, relevance, semantic specificity, timeliness, accessibility, and reproducibility. While the data is marked accordingly with a metadata item for the respective quality indicators, the metadata items are enhanced with an additional metadata item regarding reliability [14].
Results
The UMG-MeDIC infrastructure now operates based on the microservice paradigm, where each application functions independently. The microservice paradigm describes an architectural style for the development, where a large application is divided into smaller, independent elements, with their own respective realm of responsibility [12]. By detailing how the UMG-MeDIC infrastructure utilizes a microservice paradigm to independently manage metadata extraction and storage in a medical data warehouse, using an open-source framework that supports data provenance and process validation [12] research question RQ1 was adressed.
With the creation of a convergence format that integrates metadata items from various healthcare data standards, this research focused on the objective outlined in the research questions RQ2. Metadata were extracted from clinical care systems, and a generic database schema was implemented to house these metadata. The convergence format, as outlined in Bönisch et al. [13], demonstrated significant effectiveness in harmonizing metadata from disparate data sources without losing critical information. This format generated required metadata items during transformation. It ensures the retention of data integrity across formats, preserving the completeness and relevance of metadata during data transformations. Testing of the convergence format showed compatibility with the evaluated formats and the ability to adapt metadata elements from each format, facilitating interoperability.
In line with RQ3 “How can the reliability of metadata be demonstrated and what quality metrics must be met for this to be possible?“ the study defined a set of quality indicators – completeness, consistency, correctness, correspondence, relevance, semantic specificity, timeliness, accessibility, and reproducibility – to evaluate the quality of clinical metadata stored within the UMG-MeDIC [14]. These indicators were systematically applied to each metadata item, allowing for comprehensive quality assessment across diverse data structures. As a single-case study, this research leveraged the unique data environment provided by UMG-MeDIC, where documentation, direct observation, and analysis of physical artifacts were used to gather data and validate the methodologies used. The iterative process of refining the UMG-MeDIC infrastructure based on these observations resulted in a profound data integration framework.
Overall, it was demonstrated that the UMG-MeDIC could successfully manage, store, and ensure the quality of clinical metadata, thereby advancing the reliability and accessibility of clinical care data for research purposes.
Discussion
This work highlights the feasibility and effectiveness of establishing a robust metadata management infrastructure in clinical care data through the development and implementation of the UMG-MeDIC. Addressing the objectives posed by the research questions, this study provides insight into the methodologies, architectural choices, and quality measures necessary to support metadata extraction, integration, and quality assurance within the UMG-MeDIC.
It demonstrates the feasibility of implementing a microservice-based data integration center with a metadata management application that can ensure metadata reliability and interoperability. The outcomes suggest that with adequate architectural planning, metadata convergence strategies and quality assessment frameworks, institutions can advance the accessibility and reliability of clinical care data for research purposes. Further research may explore the adaptation of these approaches in multi-center collaborations to enhance data interoperability on a broader scale, potentially paving the way for an integrated health data infrastructure.
Based on the results, the quality of medical data within the UMG-MeDIC can be determined, which is followed by qualitative evaluation of data from clinical systems within the UMG for the first time.
The preparatory work for setting up the MeDIC (linking hospital systems, setting up data of data storage, creation and integration of metadata) formed the essential basis. While implementing a microservice architecture and a convergence format for metadata integration, the project not only met the specific research objectives but also demonstrated a scalable model that can be adapted to other data integration centers (DICs).
The convergence format developed within this research has broader applicability beyond the UMG-MeDIC. By standardizing metadata across different healthcare data formats (e.g., CDISC, OMOP, openEHR, and FHIR), the convergence format demonstrates how metadata items from varied sources can be consolidated without losing essential information.
Although based on a single-case study, the methodological insights gained from UMG-MeDIC’s setup are broadly applicable. By documenting the design and iterative refinement of the UMG-MeDIC infrastructure, this research provides a possible blueprint that other centers can adapt, particularly those with similar data integration challenges. The single-case approach allowed a depth of understanding that can inform generalizable practices for other DICs looking to establish or enhance their own data integration frameworks.
This research contributes to the broader field of data quality and metadata management within clinical data environments by providing a structured, modular approach to metadata integration and quality assessment. While significant literature exists on the importance of metadata for data quality [15], especially in healthcare [16], this study introduces a unique application of the microservice paradigm and a convergence format for harmonizing metadata across diverse standards. In doing so, this work addresses gaps in metadata interoperability and quality validation that have not been extensively explored in existing research.
Prior studies have underscored the importance of high-quality metadata for clinical data usability and reliability. For instance, Weiskopf et al. emphasize the necessity of complete, accurate, and accessible metadata for enhancing clinical data quality in electronic health records (EHRs), noting that metadata quality directly affects the integrity of secondary data use for research and care improvementy [17]. Kahn et al. describe a framework for assessing data quality dimensions, including completeness, consistency, and timeliness, which are integral to this study’s quality metrics [18].
Unlike previous studies, which often address quality in isolation [18], [19], this research operationalizes these indicators within the UMG-MeDIC infrastructure, allowing for a systematic, scalable quality assessment of clinical metadata.
Metadata interoperability is a persistent challenge in healthcare, where diverse data formats such as CDISC, OMOP, openEHR, and FHIR must often coexist. Previous work by Martínez-Costa et al. has highlighted the challenges of aligning these standards without information loss, proposing ontology-based approaches as a partial solution [20]. This study differentiates a convergence format that enables metadata from disparate formats to coexist within a single framework, addressing the need for a practical solution in real-world clinical settings. This format not only integrates metadata items across standards but also accommodates missing data, preserving the metadata structure.
The impact on the research field can be seen in terms of improved research quality. High-quality medical data is crucial for the accuracy and reliability of research results. In addition, faster usability can be deduced. With the availability of high-quality (meta)data, researchers can save time and resources as they no longer have to spend time checking and cleaning the data themselves.
Furthermore, interdisciplinary research can be promoted with the provision of metadata on data quality and reliability.
Finally, to discuss the limitations of the work, it must be stated that the combination of qualitative and quantitative methodology led to a high level of complexity and resource intensity. The research type of the case study led to the specificity, researched exclusively for the location UMG, although results such as the calculation of the quality factors could also be transferred to other locations.
In a next step, the visualisation and provision of the metadata reliability within the UMG-MeDIC is planned and the further development of the automated quality calculation is intended to be developed in the future.
Notes
Dissertation
The article is a summary of the dissertation entitled “Automated Metadata Transformation in a Medical Data Integration Center: Implementation of an Algorithm and Standardized Quality Analysis” at the Institute for Medical Informatics of the Medical Faculty of Georg-August-Universität Göttingen [21].
GMDS-Förderpreis 2024
The dissertation “Automated Metadata Transformation in a Medical Data Integration Center: Implementation of an Algorithm and Standardized Quality Analysis” was nominated for the GMDS-Förderpreis 2024 (GMDS Sponsorship Award for Students 2024) for the best thesis in the field of medical informatics (https://www.gmds.de/en/preise-ehrungen/gmds-foerderpreise-fuer-studierende/).
Author contributions
C.B. performed the literature searches and reviews, provided the metadata crosswalk and performed the realization of the priority classification and calculated the scoring. She established the quality factors, carried out the quality calculations and coded the Metadata Explorer.
Author‘s ORCID
Prof. Dr. Caroline Bönisch: 0000-0001-7169-6090
Competing interests
The author declares that she has no competing interests.
Funding
The author acknowledges funding, received from the German Federal Ministry of Education and Research for the project FAIRRMeDIC, grant number 01ZZ2012.
References
[1] Gaddale JR. Clinical Data Acquisition Standards Harmonization importance and benefits in clinical data management. Perspect Clin Res. 2015 Oct-Dec;6(4):179-83. DOI: 10.4103/2229-3485.167101[2] Gliklich RE, Leavy MB. Assessing Real-World Data Quality: The Application of Patient Registry Quality Criteria to Real-World Data and Real-World Evidence. Ther Innov Regul Sci. 2020 Mar;54(2):303-7. DOI: 10.1007/s43441-019-00058-6
[3] Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc. 2013 Jan 1;20(1):144-51. DOI: 10.1136/amiajnl-2011-000681
[4] Bonomi L, Jiang X. Patient ranking with temporally annotated data. J Biomed Inform. 2018 Feb;78:43-53. DOI: 10.1016/j.jbi.2017.12.007
[5] U.S. Food And Drug Administration. Real-World Evidence. 2023 Feb 5. Available from: https://www.fda.gov/science-research/science-and-research-special-topics/real-world-evidence
[6] Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJ, Groth P, Goble C, Grethe JS, Heringa J, 't Hoen PA, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016 Mar 15;3:160018. DOI: 10.1038/sdata.2016.18
[7] Baca M, editor. Introduction To Metadata. 3rd ed. Los Angeles: Getty Research Institute; 2016. (The Getty Research Institute Publications Program).
[8] Canham S, Ohmann C. A metadata schema for data objects in clinical research. Trials. 2016 Nov 24;17(1):557. DOI: 10.1186/s13063-016-1686-5
[9] Sinaci AA, Núñez-Benjumea FJ, Gencturk M, Jauer ML, Deserno T, Chronaki C, Cangioli G, Cavero-Barca C, Rodríguez-Pérez JM, Pérez-Pérez MM, Laleci Erturkmen GB, Hernández-Pérez T, Méndez-Rodríguez E, Parra-Calderón CL. From Raw Data to FAIR Data: The FAIRification Workflow for Health Research. Methods Inf Med. 2020 Jun;59(S 01):e21-e32. DOI: 10.1055/s-0040-1713684
[10] Benbasat I, Goldstein DK, Mead M. The Case Research Strategy in Studies of Information Systems. MIS Quarterly. 1987;11:369-86. DOI: 10.2307/248684
[11] Yin RK. Case Study Research, Design and Methods. Beverly Hills, CA: Sage Publications; 1984.
[12] Parciak M, Suhr M, Schmidt C, Bönisch C, Löhnhardt B, Kesztyüs D, Kesztyüs T. FAIRness through automation: development of an automated medical data integration infrastructure for FAIR health data in a maximum care university hospital. BMC Med Inform Decis Mak. 2023 May 15;23(1):94. DOI: 10.1186/s12911-023-02195-3
[13] Bönisch C, Kesztyüs D, Kesztyüs T. Harvesting metadata in clinical care: a crosswalk between FHIR, OMOP, CDISC and openEHR metadata. Sci Data. 2022 Oct 28;9(1):659. DOI: 10.1038/s41597-022-01792-7
[14] Bönisch C, Kesztyüs D, Kesztyüs T. FAIR+R: Making Clinical Data Reliable Through Qualitative Metadata. Stud Health Technol Inform. 2024 Jan 25;310:99-103. DOI: 10.3233/SHTI230935
[15] Stausberg J, Harkener S, Jenetzky E, Jersch P, Martin D, Rupp R, Schönthaler M. FAIR and Quality Assured Data – The Use Case of Trueness. Stud Health Technol Inform. 2022 Jan 14;289:25-8. DOI: 10.3233/SHTI210850
[16] Ochoa X, Duval E. Automatic evaluation of metadata quality in digital repositories. Int J Digit Libr. 2009 Aug;10:67-91. DOI: 10.1007/s00799-009-0054-4
[17] Weiskopf NG, Bakken S, Hripcsak G, Weng C. A Data Quality Assessment Guideline for Electronic Health Record Data Reuse. EGEMS (Wash DC). 2017 Sep 4;5(1):14. DOI: 10.5334/egems.218
[18] Kahn MG, Callahan TJ, Barnard J, Bauck AE, Brown J, Davidson BN, Estiri H, Goerg C, Holve E, Johnson SG, Liaw ST, Hamilton-Lopez M, Meeker D, Ong TC, Ryan P, Shang N, Weiskopf NG, Weng C, Zozus MN, Schilling L. A Harmonized Data Quality Assessment Terminology and Framework for the Secondary Use of Electronic Health Record Data. EGEMS (Wash DC). 2016 Sep 11;4(1):1244. DOI: 10.13063/2327-9214.1244
[19] Bruce TR, Hillmann DI. The continuum of metadata quality: defining, expressing, exploiting. In: Hillman D, Westbrooks E, editors. Metadata in practice. Chicago: ALA Editions; 2004. p. 238-256.
[20] Martínez-Costa M, Jiménez-Jiménez D, Dine Rabeh HA. The effect of organisational learning on interorganisational collaborations in innovation: an empirical study in SMEs. Knowledge Management Research & Practice. 2018;17(2):137-50. DOI: 10.1080/14778238.2018.1538601
[21] Bönisch C. Automated Metadata Transformation in a Medical Data Integration Center: Implementation of an Algorithm and Standardized Quality Analysis [Dissertation]. Göttingen: Georg-August-Universität Göttingen; 2023. DOI: 10.53846/goediss-10167




